Figure 3. Microbial-metabolites and diversity as triggers for liver cancer progression.
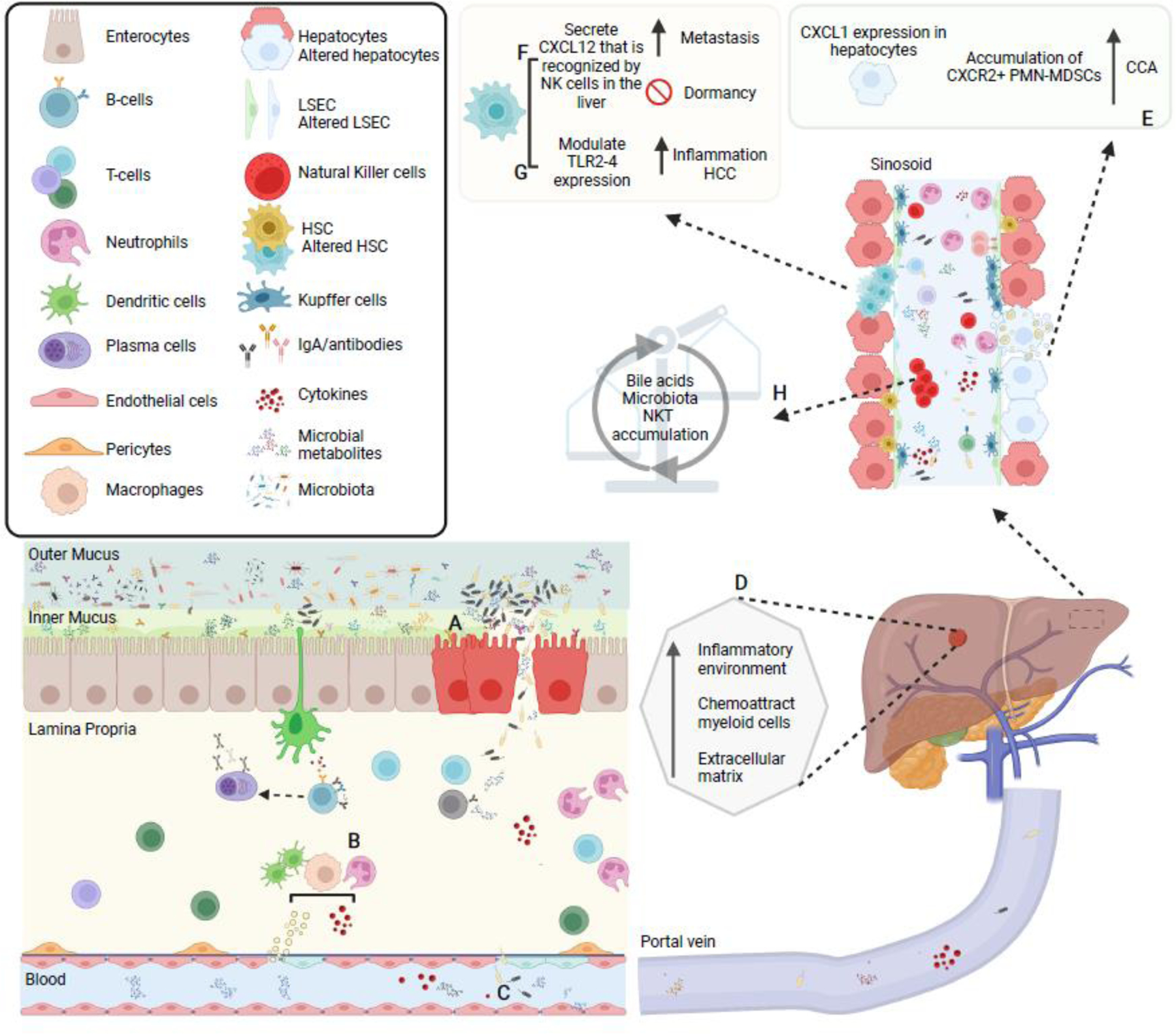
(A) Some bacteria may penetrate the mucus barrier or participate to its degradation, in some cases leading to breaching or disruption of the epithelial barrier, thus allowing (B) direct contact between microbes and microbial-metabolites with immune cells induce pro-inflammatory cytokines production and systemic dissemination. Disruption of the mucus/epithelial barriers may also facilitate (C) translocation of bacteria to the liver (D) where creates a favorable niche for cancer cell seeding. Hence, influx of previously compartmentalized bacteria and microbial products influence local liver cells’ gene expression. For example: (E) hepatocytes may express CXCR1 and induce an accumulation of CRCX2+ polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) creating an immunosuppressive environment to promote cholangiocarcinoma (CCA); (F) activated hepatic stellate cells that exert multiple functions for HCC and cancer metastasis may disrupt the function of NK cells in the liver through CXCL12-CXCR4 interactions, altering NK-cell mediated immunity and promoting breast metastasis into the liver; and (G) other liver immune cells may be activated via LPS-TLR4 or DCA-TLR2 modulation and induce inflammation favoring HCC generation. On the other hand, (H) liver produced bile acids after been modified by gut microbiota may activate a chemokine-dependent accumulation of hepatic NKT cells in the liver, controlling tumor growth.
