Figure 4.
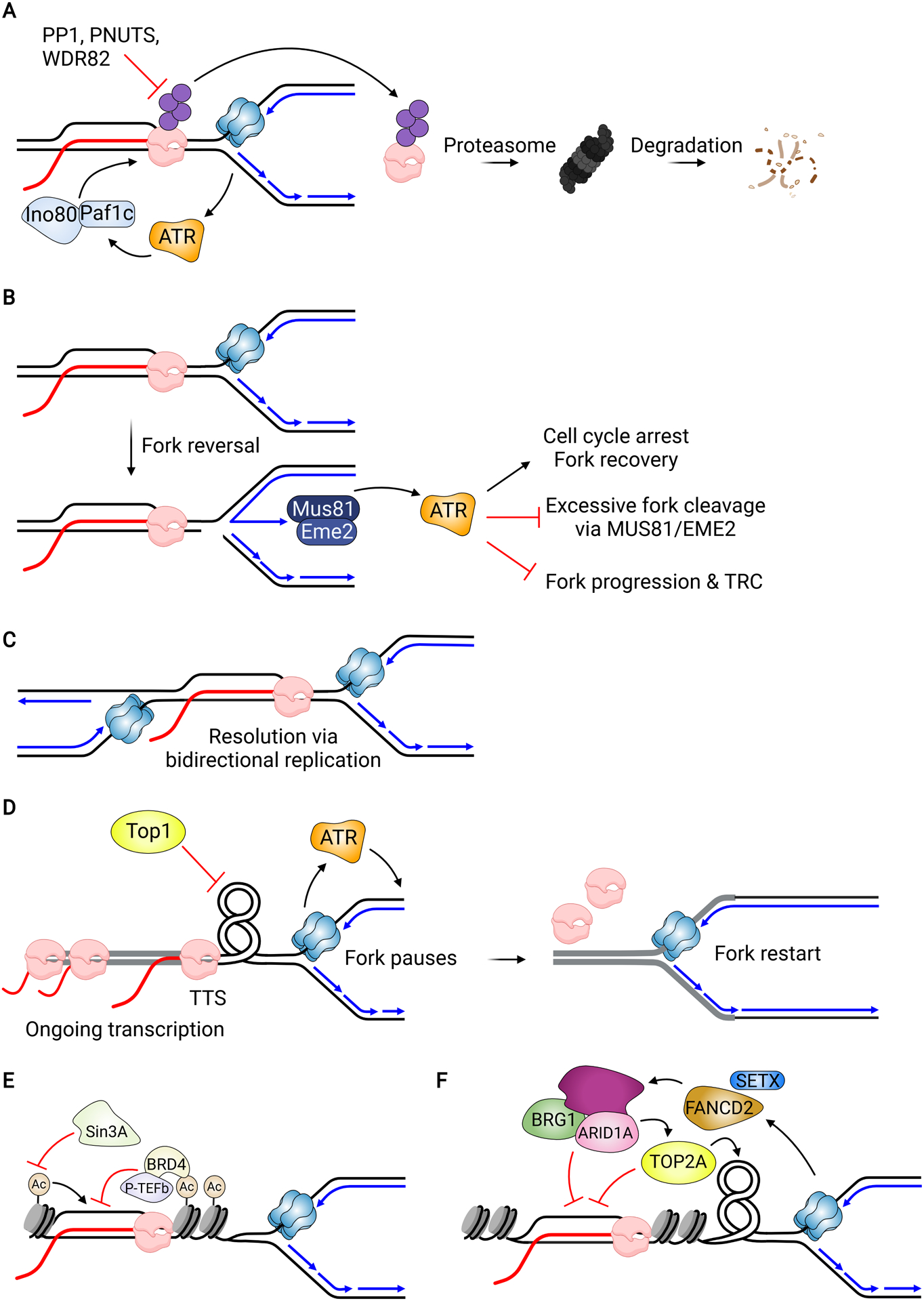
Potential Mechanisms for Resolution of R-loop Induced Transcription-Replication Conflicts. (A) In budding yeast, RNAP removal and proteasomal degradation can be initiated by activation of ATR and mediated by Paf1c and Ino80. RNAP phosphorylation also promotes its degradation, which is counteracted by the protein phosphatase PP1 and its regulatory subunit PNUTS and WDR82. (B) ATR activation at stalled forks is dependent upon MUS81-EME2-mediated fork cleavage, which is needed to generate ssDNA. Activation prevents additional and excessive fork cleavage by MUS81 and suppresses TRC formation. Other characterized functions of ATR may also promote fork recovery. (C) A fork stalled at an HO conflict can be rescued by a fork approaching the R-loop from the CD orientation. (D) Fork pausing at the 3’-end of a gene (highlighted in gray) located near an origin allows time for transcription to complete before the fork progresses into the gene. The converging fork and transcription lead to torsional stress that is relieved by TOP1. Pausing also activates ATR, which may reinforce the slowdown by inhibiting elongation. (E) Histone deacetylation suppresses R-loop formation in diverse ways, preventing genome instability. (F) SWI/SNF complexes including the subcomplex containing ARID1A act in a pathway with FANCD2 and SETX to suppress R-loop formation and TRCs. ARID1A recruits TOP2A to forks to relieve torsional stress.
