Figure 5.
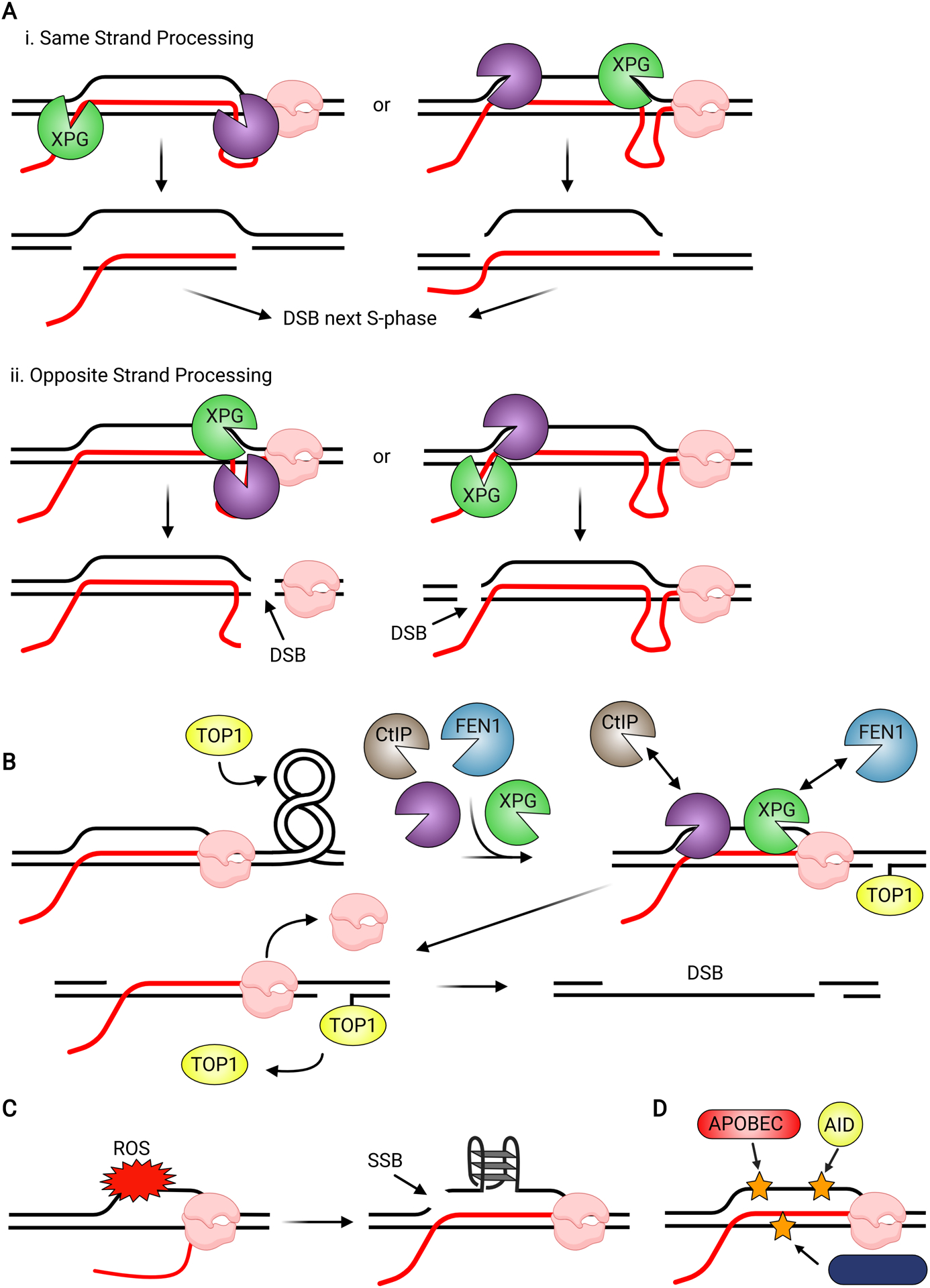
Potential Mechanisms of Replication-Independent R-loop-mediated DNA Damage. (A) The TC-NER nucleases XPG and XPF initiate R-loop processing by cleaving the 5’ or 3’-flap of the R-loop-template DNA junction, respectively. Cleavage may occur on either the template strand, excising the hybrid, or on the non-template strand, excising the displaced ssDNA to generate a single-strand break. Conversely, XPG and XPF may cleave opposing DNA strands of the R-loop, generating a DSB either 5’ or 3’ to the R-loop. (B) R-loop processing by several nucleases and TOP1-mediated DNA nicking in response to camptothecin generate two SSBs on opposing DNA strands, which mimic a DSB. (C) The displaced ssDNA of a transcription bubble is more accessible to damage from reactive oxygen species (ROS). ROS-generated SSBs in the displaced ssDNA promote unscheduled R-loops and G4s. (D) AID and APOBEC proteins can convert cytosine to uracil in the displaced ssDNA. Downstream processing of uracil results in an abasic site, which can cause SSBs. Additionally, the RNA editor ADAR has some activity towards the DNA strand of the hybrid. Base modifications are represented by stars.
