Figure 4 |. KKR region in GluN2a and GluN2b is required for the direct binding to the EE3 motif in TRPM2.
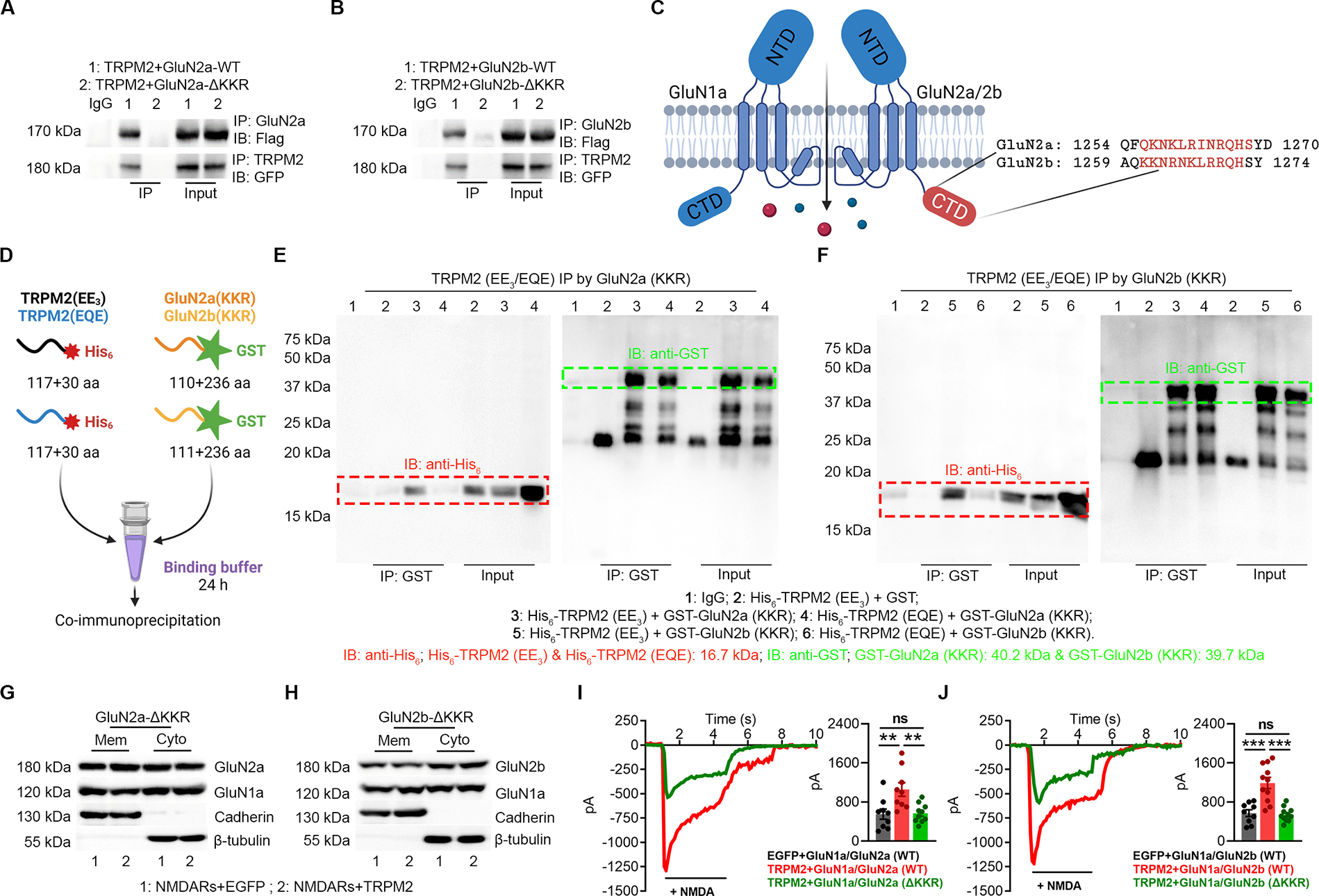
(A-B) Co-IP of KKR region deleted GluN2a (GluN2a-ΔKKR, A) and GluN2b (GluN2b-ΔKKR, B) with TRPM2. TRPM2 is flag-tagged and GluN2a/GluN2b is GFP-tagged. IP using anti-GluN2a/b and IB with anti-Flag (upper). IP using anti-TRPM2 and IB using anti-GFP (lower).
(C) Structure of GluN2a and GluN2b. The KKR domain (red label indicates the deleted sequence (ΔKKR)) is localized within the C terminal domain (CTD).
(D), Schematic diagram of the in vitro binding assay. EE3 and EQE containing fragments were labelled by a N-terminal His6 tag and KKR containing fragments were labelled by a N-terminal GST tag.
(E-F) Co-IP of EE3 and EQE containing fragments with KKR containing fragments from GluN2a (E) and GluN2b (F). IP using anti-GST and IB using anti-His6.
(G-H) Surface expression of NMDARs in HEK-293T cells co-transfected with TRPM2 and GluN2a-ΔKKR (G) and GluN2b-ΔKKR (H).
(I, J) NMDAR current recording in HEK-293T cells co-transfected with TRPM2 and GluN2a-ΔKKR (I) and GluN2b-ΔKKR (J).
(**, p<0.01; ***, p < 0.001; ANOVA, Bonferroni’s test; mean ± SEM)
