Figure 7. Knockdown of the MTMR5-MTMR2 axis enhances the proteolytic clearance of Dendra2.
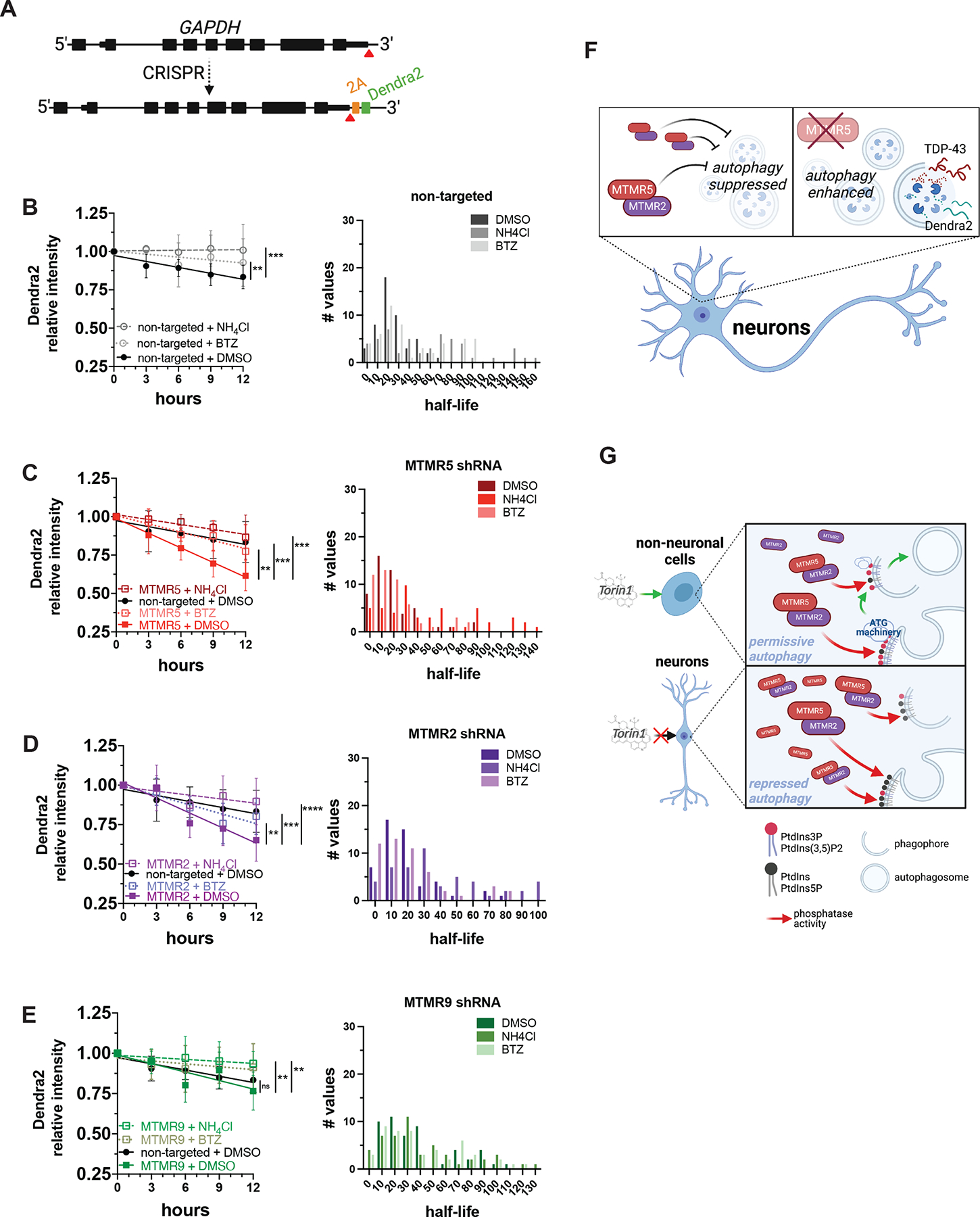
(A) Strategy for inserting Dendra2 at the native GAPDH locus using CRISPR/Cas9. 2A, self-cleaving peptide. (B–E) Dendra2 fluorescence measured in GAPDH-2A-Dendra2 iNeurons after knockdown of each indicated MTMR by transduction with non-targeted (B), SBF1 (C), MTMR2 (D), or MTMR9 (E) shRNA lentivirus and after the indicated drug treatments (left panels), and histogram plot of the half-lives of each measured iNeuron (right panels). Data are represented at each time point as mean ± SD; **p<0.01; ***p<0.001, one-way ANCOVA. (F) Schematic summary of MTMR5 influence on the proteolysis of autophagy substrates. In neurons, autophagy induction is suppressed by native levels of MTMR5 (left), but opposing MTMR5 activity (e.g., through shRNA-mediated knockdown) disinhibits autophagy and accelerates autophagic degradation of substrates, such as TDP-43 and Dendra2 (right). (G) Working model of cell type-specific regulation of autophagy. In non-neuronal cells (left), autophagy operates under permissive conditions due to relatively lower levels of MTMR5, enabling sufficient levels of PtdIns3P to recruit autophagy-related protein complexes (ATG machinery) necessary for autophagosome biogenesis and in response to stimuli (e.g., Torin1). However, in neurons (right), higher levels of MTMR5 impair autophagy induction by potentiating MTMR2, depleting PtdIns3P scaffolds necessary for assembling ATG machinery, and leading to a repressed state of autophagy induction (right panel). See also Figure S6.
