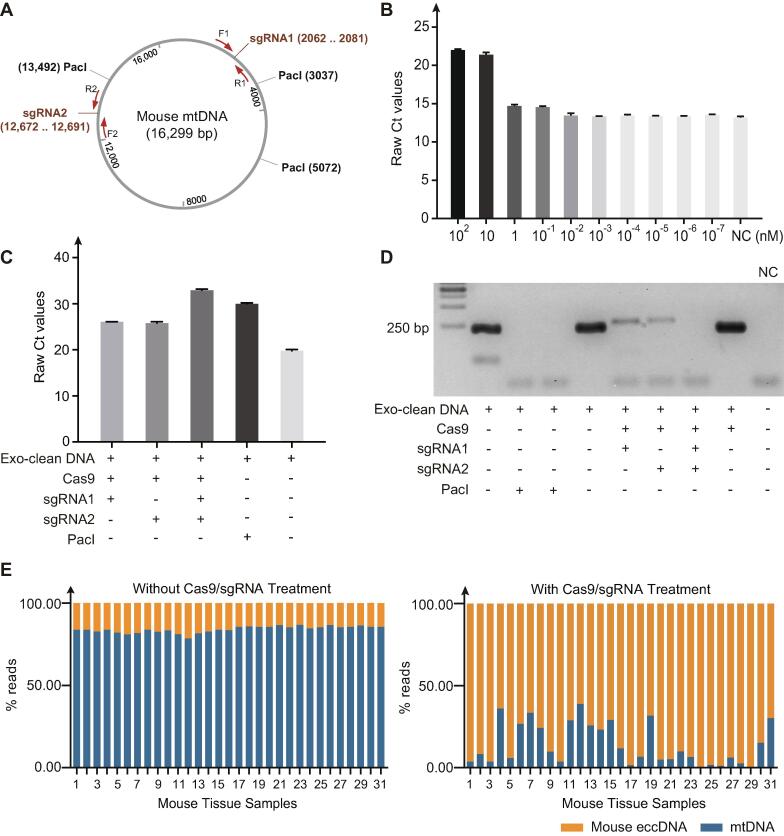
Fig. 2.
Removal of mtDNA in mouse tissues. (A) Cleavage site of the two sgRNAs on mouse mtDNA (Table 1). (B) Quantitative real-time PCR (qRT-PCR) confirmation of the cutting efficiency of different Cas9/sgRNA concentrations (primer F1 and R1). Cas9/sgRNA were diluted at the concentration of 102 nM, 10 nM, 1 nM, 10−1 nM, 10−2 nM, 10−3 nM, 10−4 nM, 10−5 nM, 10−6 nM, 10−7 nM, and then subjected to the cleavage reaction of mtDNA. Followed by cleavage efficiency detection with qPCR. All the assays were performed three times. Error bars: mean ± SD of three independent tests. (C) Quantitative real-time PCR (qRT-PCR) confirmation of the CRISPR-Cas9 cleavage efficiency (primer F2 and R2). DNA digested with restriction enzyme PacI was used as a positive control; DNA only treated with cas9 protein was used as a negative control. (D) PCR validation of CRISPR-Cas9 treated DNA (primer F2 and R2). Cas9/sgRNA were diluted at the concentration of 10 nM, and then subjected to the cleavage reaction of mtDNA using sgRNA1, sgRNA2, and both two sgRNAs respectively. Followed by cleavage efficiency detection with PCR. DNA digested only with ExonucleaseV (Exo-clean DNA) was used as a negative control; DNA digested with restriction enzyme PacI was used as a positive control (two technical replicates); H2O was used as non-template control. (E) MtDNA reads mapping ratio with and without CRISPR-Cas9 treatment. DNA was extracted from 9 different mice tissues (Table S1) including 3 pancreas, 2 subcutaneous adipose tissue, 5 hippocampus, 1 visceral adipose tissue, 7 cortex, 5 skeletal muscle, 2 skin, 2 whole skin with hair and 4 liver samples, followed by linear DNA removal, rolling circle amplification, library preparation and next-generation sequencing.