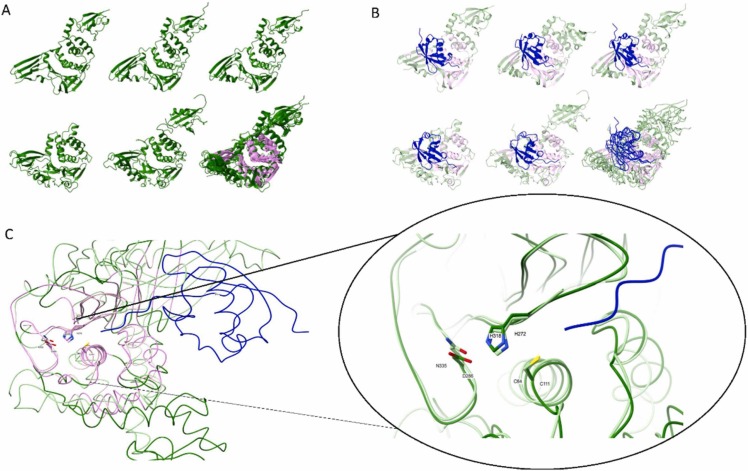
Fig. 3.
Structural alignment of viral PLpro and select USP enzymes. A. Ribbon illustration of apoenzyme 3D structures: i) MERS-PLpro (pdb:4rna), ii) SARS-CoV-PLpro (pdb:2fe8), iii) SARS-CoV2-PLpro (pdb:6wrh), iv) hUSP18 (AF-Q9UMW8-F1; [130]), and v) hUSP14 (pdb:6iik+1wgg). PDB entries were taken from https://www.rcsb.org/ and AlphaFold models from https://alphafold.ebi.ac.uk/. A full 3D structure of hUSP14 was generated by superimposing the USP domain (6iik) and N-terminal UBL domain (1wgg) onto the PLpro structure. A model of hUSP18 was generated by Alphafold [130]. vi) Superimposition of all five enzymes highlights structurally equivalent residues shared by all five enzymes (i.e. the common core shown in pink) calculated by mTM-align [131]. B. Ribbon illustration of PLpro and USP 3D structures in complex with substrate, either Ub (hUSP14) or proximal domain of ISG15: i) MERS-PLpro (pdb:5w8u), ii) SARS-CoV-PLpro (pdb:5tl7), iii) SARS-CoV2-PLpro (pdb:6xa9), iv) hUSP18 (AlphaFold model:AF-Q9UMW8-F1 +PDB:2hj8), and v) hUSP14 (PDB:2ayo+1wgg). Full 3D structure of Ub-bound hUSP14 was generated by the superimposition of 2ayo for the Ub-aldehyde domain bound to the USP domain and 1wgg for the N-terminal UBL domain of hUSP14, onto the structure of PLpro. A model of ISG15-bound hUSP18 was generated by Alphafold [130] for hUSP18 and the solution structure of the proximal domain of hISG15 (PDB:2hj8) superimposed on complexed mUSP18-mISG15 (PDB:5CHV). vi) Superimposition of substrate-bound enzymes defines the common core. Structurally equivalent residues of all five enzymes are shown in pink as computed by mTM-align [131]. C. Superimposition of the catalytic core of hUSP18 and ISG15-bound SARS-CoV2-PLpro (as determined in panel A). The shared residues of both these proteases calculated by mTM-align[131] are shown in pink; left. The catalytic triad residues overlay precisely in the active site; right. The narrow cleft leading to this active site restricts access to substrates with flexible stretches (shown in blue). Cleavage is limited to the presence of Glycine at P1 and P2 sites. Images were generated using ChimeraX [132].