Abstract
A pre-eminent subtype of lung carcinoma, Non-small cell lung cancer accounts for paramount causes of cancer-associated mortality worldwide. Undeterred by the endeavour in the treatment strategies, the overall cure and survival rates for NSCLC remain substandard, particularly in metastatic diseases. Moreover, the emergence of resistance to classic anticancer drugs further deteriorates the situation. These demanding circumstances culminate the need of extended and revamped research for the establishment of upcoming generation cancer therapeutics. Drug repositioning introduces an affordable and efficient strategy to discover novel drug action, especially when integrated with recent systems biology driven stratagem. This review illustrates the trendsetting approaches in repurposing along with their numerous success stories with an emphasize on the NSCLC therapeutics. Indeed, these novel hits, in combination with conventional anticancer agents, will ideally make their way the clinics and strengthen the therapeutic arsenal to combat drug resistance in the near future.
Keywords: Lung cancer, Drug resistance, Drug Discovery, Drug repurposing, Repositioned hits
1. Lung cancer biology
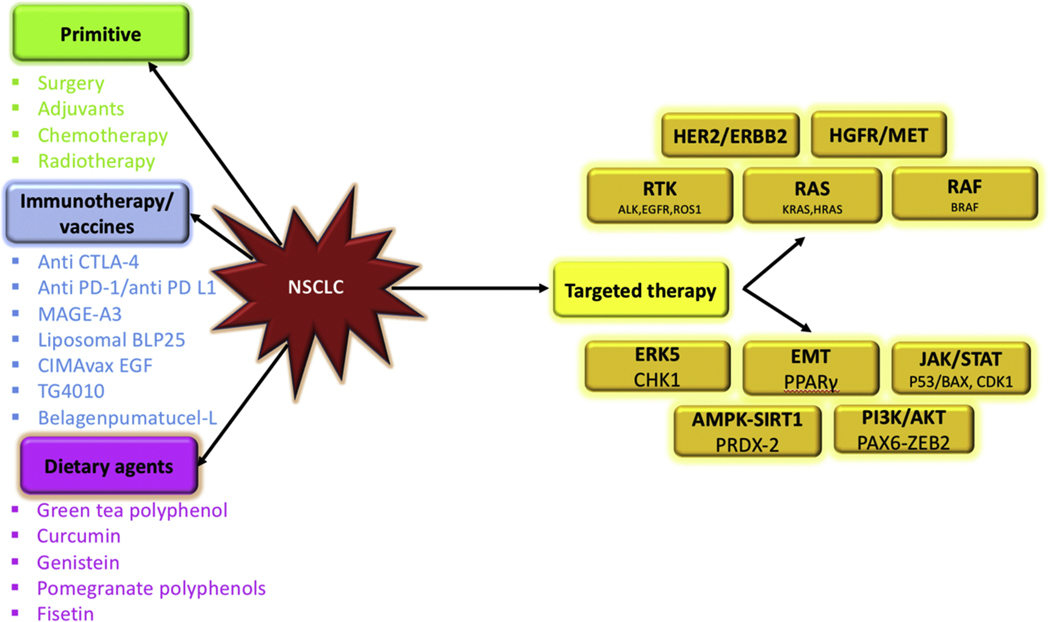
Lung cancer harbours a complicated molecular basis portraying diverse heterogeneity. Hence, it is obligatory to comprehend the technicalities in order to invent targeted therapies. Lung cancer is majorly bifurcated into non-small cell lung cancer (NSCLC) represented by 85% of the case and the rest is attributed to small cell lung cancer (SCLC). Histologically, NSCLC is withal systematized into large-cell carcinoma, squamous-cell carcinoma and adenocarcinoma, [1]. Literature evidences support that squamous-cell lung carcinoma emanates at the main bronchi and forges ahead towards the carina. With a slight different, adenocarcinoma springs among the peripheral bronchi. Moreover, large-cell carcinoma depicts exiguity of either squamous or classic glandular morphology. Finally, small-cell lung cancer originates from the hormonal cells, promulgates into regional lymph nodes and submucosal lymphatic vessel in the absence of a bronchial invasion. Under normal conditions, several proteins work in an intricate manner in order to establish homeostasis and normal cell proliferation. Lung cancer develops through an involuted mechanism which comprises of several genetic and epigenetic alterations, specifically upregulation of oncogenes and downregulation of tumor suppressor genes [2]. Subsequent to the initiation of primary cancer, perpetuating amassment of aberrant genetic transformation, procured in the course of clonal expansion, predisposes the mechanism of metastasis, tumor invasion and resistance to cancer medication. A few tumor suppressor genes and oncogenes have been characterized which are found to be altered in NSCLC. The RTK pathway includes the oncogenes namely, ALK, MET, DDR2, EGFR, ERBB2/3, FGFR1 [3]. The RAS pathway is marked by the genes HRAS and KRAS [4]. Similarly, the RAF pathway involves BRAF gene [5]. A few of these proteins are depicted in Fig. 1. Apart from this, enhanced telomerase activity, which maintains the telomere length through incessant synthesis of telomeres and lengthing of enduring telomeres, contributes towards the cellular immortality [6]. Additionally, the tumor microenvironment plays a very crucial role to support the tumor growth. The complex interaction of the cancer-associated fibroblast, stromal cells, immune cells and stem cells constitute major hallmarks of cancer [7]. This microenvironment gets transmuted in a manner so as to foster tumor growth and suppress the host’s immune system. The altered environment is marked by effectual secretion of sundry growth factors like vascular endothelial growth factor (VEGF) and granulocyte-macrophage colony stimulating factor (GM-CSF) and by the loss of antigen variants as well as MHC class I molecules. The immune cells are functionally vitiated, and the newly infiltrating immune cells get triggered, causing a disconcerted phenotype [8]. Thus, lung cancer is equipped with multifarious targets and understanding the molecular biology of lung cancer will help us to formulate targeted therapies.
Fig. 1.
Traditional and rationale strategies for the treatment of NSCLC.
2. Treatment methods
There are ample amount of treatment options available for cancer. They can be majorly categorized into primitive, immunotherapy, existing and advanced targeted therapy. The primitive methods focus on either its physical removal or destroy it with the help of radiation. A few routine procedures which have been implemented for lung cancer are surgery, adjuvants, chemotherapy and radiotherapy. Either the tumor is physically removed by surgery or adjuvant therapy takes charge. This refers to the subsequent treatments viz chemotherapy, radiotherapy or targeted therapy [9]. Advances in scientific research has suggested that the immune system of the body is a very attractive option for cancer treatment [10]. This is the in-built surveillance network, which protects the body from diseases by keeping a check on the foreign particles and maintain harmony among various life processes. For long, it was considered that lung cancer is non-immunogenic. But lately, evidences depict the immune system evasion by lung cancer [10]. There is loss of major histocompatibility complex (MHC), secretion of immunosuppressive cytokines, and expression of molecules that prevent the activation of T cells. All these have paved way for immunotherapeutic. This futuristic therapy aims to elicit the immune-mediated destruction of cancer cells [11–13]. Presently, there are monoclonal antibodies, immunomodulators, therapeutic vaccines, autologous cellular therapies, and more [11–13]. Recently, checkpoint inhibitors have been discovered that target programmed death-1 (PD-1) pathway and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) [10]. Apart from this, a few vaccines have been introduced as well [14]. An immunotherapeutic NSCLC marker, MAGE-A3 antigen is composed of a recombinant fusion protein (MAGE-A3 and protein D of Hemophilus influenza) in combination with an immune-enhancing adjuvant. These help to ameliorate the immune system of the body and prepares it to attack the tumor. A few other well-known vaccines are liposomal BLP25, CIMAvax EGF, TG4010, belagenpumatucel-L [14]. Since all these futuristic approaches have paved the way for modern drug development, dietary agents are being ignored. Though they are not as effective, but they are natural and devoid of all the side effects [15]. Recent literature suggests that dietary modifications might reduce lung cancer incidences [16]. It has been found that green tea polyphenols upregulate p53 expression, genistein, curcumin and fisetin inhibits cell proliferation and promotes several other antitumor activities [17]. Components of pomegranate polyphenols like Punicalagin causes inhibition of DNA adducts and has anti-proliferative effects [17]. Finally, targeted therapy always has a place reserved in drug discovery. US Food and Drug Administration (FDA) has approved various medications for the treatment of NSCLC. For the past few years, these remedies focussed on oncogenic EGFR (epidermal growth factor receptor) mutations, ROS1 (proto-oncogene receptor tyrosine kinase) and ALK (anaplastic lymphoma kinase) fusion, BRAF and KRAS mutations, HGFR/MET (hepatocyte growth factor receptor) alterations and HER2/ERBB2 (human epidermal growth factor receptor 2) mutations [18–20]. EGFR triggers the cell growth signalling pathways, hence their oncogenic transformation leads to uncontrolled cell growth. Presently, there are four FDA-approved EGFR TKIs which are under clinical use. The first, second and third generation inhibitors include erlotinib gefitinib, afatinib and osimertinib respectively [18,21]. Similarly, ALK gene exists as a fused product with echinoderm microtubule-associated protein-like protein 4 (EML4) and enhances malignant growth and proliferation. Crizotinib was the first generation ALK inhibitor, followed by ceritinib, alectinib and brigatinib have also been approved by FDA to target ALK fusion driven NSCLC [22]. Notably, ROS1 gene rearrangements have been found to occur in a few NSCLC cases. These fusion brings together the intact ROS1 kinase domain with a range of partners, viz. CD74, to promote constitutive ROS1 kinase activity [23]. Crizotinib has addressed a few of these cases. Moreover, mutations in the BRAF gene also occur in a few lung adenocarcinoma cases. BRAF-V600E mutations induce constitutive activation of BRAF in its monomeric form, activating the downstream MEK-ERK signalling [24]. Lately, a combination of dabrafenib and trametinib have been approved for the treatment [18] Furthermore, there is a tumefying spectrum of oncogenic driver alterations, which include the MET, KRAS and HER2 mutations. Crizotinib and cabozantinib have been utilised for targeting MET alterations. Besides, KRAS signaling is targeted by the cyclin-dependent kinase 4/6 (CDK4/6) which is in-turn inhibited by abemaciclib. Lastly, HER2 has been targeted with the help of the monoclonal antibody trastuzumab in combination with chemotherapy. Shedding light upon the most recent techniques it has been established that NSCLC propagates with the help of very intricate network which involves ambiguous modifications in umpteen proteins. Neoteric literature survey provides powerful insight into these mechanisms. Nitrosylation of Peroxiredoxin-2 (Prdx-2) facilitates the apoptosis of NSCLC cells via the AMPK-SIRT1 pathway [25]. Further, UV-irradiated apoptotic cancer cells interact with macrophages in order to furnish an anti-tumor microenvironment. This interaction leads to the formation of Peroxisome proliferator activated receptor gamma (PPARγ), which is a potent PTEN transcription factor. PTEN downregulates AKT and hence curbs the epithelial to mesenchymal transition (EMT), thus inhibiting lung metastasis [26]. Moreover, increased expression of p53 and Bax, with the help of a natural extract, caused the reduction in the level of JAK2 and phosphorylated STAT3, thus prevents metastasis by inducing apoptosis in cancer cells [27]. Furthermore, Cyclin dependent kinase 1 (CDK1), which is a key determinant of mitotic progression, when bound directly to iron, promotes JAK1 phosphorylation and activates STAT3 signaling, and hence tumor formation [28]. CDK1 knockdown and DFO (iron chelator) suppresses tumorigenicity [28]. Besides, Extracellular signal-regulated kinase 5 (ERK5) is an indicator of radioresistance of cancer cells. ERK5 triggers Checkpoint kinase 1 (Chk1) thus supplementing the DNA repair of the cancer cells. ERK knockdown enhances G2/M cell cycle arrest and apoptosis, thus exhibiting anticancer activity [29]. Yet another, Paired-box 6 (PAX 6) is an oncogene, which regulated the transcriptional activity of zinc finger E-box binding protein 2 (ZEB2). PAX6-ZEB2 promotes metastasis by mediating E-cadherin down-regulation through PI3K/AKT pathway in NSCLC. PAX6 knockdown and PI3K-AKT inhibitor restricts cell migration and promotes anti-tumor activity [30]. All the treatment strategies have been summarised in Fig. 1.
3. Channelization towards drug repurposing
Despite the advancement in the treatment strategies and enhanced knowledge of cancer heterogeneity, translation of these benefits into therapeutic advances has been fewer than expected. Importantly, limited success with current therapies in advanced stages have driven huge investments into drug development. This appetency for more effective anti-cancer drugs has sparked a growing interest for drug repurposing. Thus, we have curated the details of repurposed candidate particularly for the treatment of NSCLC by extracting documents from PubMed central and Elsevier journals. Emphatically, our anatomization will bestow valuable information for the scientific community to accelerate NSCLC drug discovery.
4. Drug repurposing approaches
Although, the number of drug repositioning approaches have dramatically escalated in the recent years, they mostly fit into three basic units viz. (i) drug (ii) target and (iii) disease/therapy-oriented. On the basis of procurable information of disease and drugs we have tried to obtain meaningful interpretations for repurposing hypotheses. Here, we have highlighted the success stories of drug repurposing in lung cancer with respect to six major strategies.
4.1. Knowledge-based drug repurposing
Knowledge-based drug repurposing basically involves unveiling the novel therapeutic uses of the pre-existing medicaments with the support of the available information on drugs and their targets networks, chemical configuration, approval labels from FDA, clinical trial data and metabolic pathways into drug repurposing studies. This method grabs the lime light on account of the monumental amount of documentation that is already available, this accurate channelization and intellectual conclusion will pave way to drug repurposing. The following representative, Metformin, constitutes a magnificent illustration of the outcome of knowledge-based drug repurposing approach for NSCLC. It stands distinguished as the unrivalled exemplification of cancer drug-repurposing issued with the help of knowledge-based approach. A study was conducted by amalgamating two massive electronic health records which suggested that metformin was associated with decreased cancer mortality either by insulin-dependent or independent mechanism [31,32]. Recent evidence highlighted that its anti-tumor effect was on account of the inhibition of the mTOR signaling pathway by activating AMPK regulator and p53 [33]. Preclinical analyses have also highlighted that metformin can harmonize with chemotherapeutics like taxane and platinum, aggrandize anti-cancer potency of tyrosine kinase inhibitors and triumph over resistance to same, and function as a radio sensitizing agent [34]. Ex post facto and epidemiological studies have syndicated metformin’s use with declining lung cancer prevalence, meliorated overall survival (OS) as well as concertion with TKIs [35–37]. Hydroxychloroquine (HCQ) is an alternative illustration of the knowledge-based repurposing approach, and stands as one of the promising samples which also is an established antimalarial agent. Scientific evidence supports the HCQ has been used in cancer therapy, particularly to upgrade the quality of life of NSCLC valetudinarian. It functions as a chemo-activator and immune regulator to supplement the therapeutic effects. A study demonstrated that HCQ decreases lysosomal acidification by emancipating chemotherapeutic drugs from the lysosome to the cytoplasm or nucleus, ramifying into intensified cytotoxicity [38,39]. Moreover, HCQ does not affect cell viability, exclusively, but at higher concentrations, while 5 μM is sufficient to sensitize tumor cells [40]. The other repurposed hits emerged from this strategy were given in Table 1 [41–52].
Table 1.
Details of repurposing opportunities for the treatment of NSCLC.
| S. No | Drug Name | Original indication | Remarks | Type of Repurposing Approach | References |
|---|---|---|---|---|---|
|
| |||||
| Metformin | Diabetes | Clinical Trials Phase II | Knowledge based approach | [31,32] | |
| Hydroxychloroquine | Malaria | Clinical Trials Phase II - Ongoing | Knowledge based approach | [38] | |
| Verapamil | Antihypertensive | Randomized Clinical study | Knowledge based approach | [41] | |
| Clarithromycin | Bacterial infections | In vitro and in vivo | Knowledge based approach | [42] | |
| Dihydroartemisinin (DHA) | Malaria | In vitro and in vivo | Knowledge based approach | [43] | |
| Pirfenidone | Idiopathic pulmonary fibrosis | In vitro and in vivo | Knowledge based approach | [44] | |
| Disulfiram | Treatment of chronic alcoholism | Clinical Trials Phase III - Completed | Knowledge based approach | [45] | |
| Ruxolitinib | Psoriasis | Clinical Trials Phase I - Completed (with Afatinib) Phase II- Completed. (With erlotinib) |
Knowledge based approach | [46] | |
| Phenformin | Diabetes | In vitro and In vivo | Knowledge based approach | [47] | |
| Nelfinavir | HIV | Clinical Trials Phase II - Ongoing | Knowledge based approach | [48] | |
| Ibrutinib | Chronic lymphocytic leukemia | Clinical Trials Phase II - Ongoing | Knowledge based approach | [49,50] | |
| Tigecycline | Antibiotic | In vitro and in vivo studies | Knowledge based approach | [51,52] | |
| Pimozide | Antipsychotic | In vitro | Signature based approach (Disease) | [56] | |
| Trifluoperazine | Antipsychotic drug | In vitro and in vivo studies | Signature based approach (Gene) | [57] | |
| Bisphosphonates | Osteoporosis and metastatic bone disease | In vivo | Signature based approach (Drug) | [58] | |
| Ritonavir | HIV | In vitro | Signature based approach (Gene) | [59] | |
| Carglumic acid | Hyperammonemia | In vitro and in vivo | Pathway based approach | [61] | |
| Simvastatin | High cholesterol | Clinical Trials Phase II - Ongoing | Pathway based approach | [62] | |
| Celecoxib | Fever and Pain control | Clinical Trials Phase I - Completed | Pathway based approach | [63] | |
| Apricoxib | Fever and Pain control | Phase III | Pathway based approach | [63] | |
| Rofecoxib | Fever and Pain control | Phase II | Pathway based approach | [63] | |
| Lovastatin | Hypercholesterolemia | In vitro | Pathway based approach | [64] | |
| Fluphenazine | Anti-psychotic | in vivo and in vitro validation | Network Based (Protein-Protein network based approach with cMap) | [65] | |
| Perphenazine | Schizophrenia | in vivo and in vitro validation | Network Based (Protein-Protein network based approach with cMap) | [65] | |
| Mefloquine | Malaria | in vivo and in vitro validation | Network Based (Protein-Protein network based approach with cMap) | [65] | |
| Auranofin | Rheumatoid arthritis | In vitro and in vivo studies | Mechanism of action | [67,68] | |
| Nitroglycerin | Coronary vasodilator | Clinical Trials Phase II | Mechanism of action | [69] | |
| Clobetasol propionate | Skin disorders | Only in vitro and in vivo studies | Target based approach | [70] | |
| Albendazole | Anti-helminthic | In vitro | Target based approach | [73] | |
| Mebendazole | Anti-helminthic | In vitro | Target based approach | [73] | |
| Cabozatinib | Medullary thyroid cancer and renal cell carcinoma | In vitro | Target based approach | [75] | |
| Sertraline | Antidepressant | In vitro and in vivo | Network Based approach (Integration of the drug-gene interaction (DGI) and the gene-disease association network (GDN)) | [91] | |
| Bosutinib | Chronic myelogenous leukemia | Only in vitro validation | Integrative systems biology approach | [92] | |
| Bezafibrate | Hyperlipidaemia | In vitro and in vivo | Multiple approach (expression-based in silico screening + CMap network analysis) | [93] | |
| Mepacrine (Quinacrine) | Antiparasitic | Phase I - Completed | Gene expression analysis + CMap + Machine learning approach | [94] | |
| Itraconazole | Anti-fungal | Phase II studies – ongoing | Activity based repurposing approach and pathway based approach | [95,96] | |
| Potassium Antimonyl Tartrate (PAT) | Antiparasitic Drug | In vitro | Blinded search or screening approach | [97] | |
| Bortezomib | Refractory multiple myeloma | Clinical Trials Phase II - Ongoing | Blinded search or screening approach | [98,99] | |
| Ouabain | Cardiovascular disease | In vitro | Blinded search or screening approach | [100] | |
| Rifabutin | Tuberculosis | Only in vitro and in vivo studies | Blinded search or screening approach | [101] | |
| Niclosamide | Anti-helminthic | In vitro | Blinded search or screening approach | [102] | |
4.2. Signature-based drug repurposing
The comparison between drug and disease gene expression profiles utilize the gene signatures introducing a novel method designated as ‘signature reversion’ [53,54]. A circumstantial map concatenating disease and drug actions can be constructed with the help of gene expression based methods [55]. Predominantly on the basis of transcriptome data, it was victoriously manipulated to ascertain drug repositioning opportunities in a wide range of therapeutics areas, especially in the field of oncology and rare diseases.
An antipsychotic drug, Pimozide, constitutes a striking example that was identified as lung cancer therapeutic in the context of signature-based repurposing strategy. The growth retardation effects of pimozide were studied in four cell lines of lung cancer, all of which overexpressed HCC4006, CALM1, H460, A549, and H1437. Fortunately, the study demonstrated that pimozide showed significant anticancer activity and hence can be used to treat lung cancer. However, the contrivance of its activity remains unascertained and materializes to be CALM1-independent [56]. Furthermore, another antipsychotic agent, Trifluoperazine, stands as an interesting success story of gene-signature based repurposing approach. It was postulated that trifluoperazine impedes tumor growth and overcomes drug resistance by exerting anti-CSC effects. Trifluoperazine obstructs Wnt/β-catenin signaling in gefitinib-resistant lung cancer spheroids. The coalescence of trifluoperazine with either cisplatin or gefitinib has also been reported to overcome drug resistance in lung CSCs [57]. Moreover, Bisphosphonates are a fascinating example from signature-based approach, as well. These group of drugs are the mainstay of therapy worldwide for osteoporosis and skeletal metastasis. However, the cell free in vitro assays with N-containing bisphosphonates established that they minimize tumor progression in certain subgroups of patients. It downregulates the signaling pathways by blocking kinase domain of HER1/2. Thus, it could possibly be used for HER family associated cancer drug repurposing [58]. Ritonavir, originally used as HIV Protease inhibitor, also of interest in lung adenocarcinoma therapeutics [59].
4.3. Pathway and network-based methods
The disease-specific pathway and network based approach is yet another possibility which combines disease “omics” data, metabolic pathways and protein interaction networks [60].
Carglumic acid (Carbaglu; Orphan Europe) one of the remarkable examples, was discovered by pathway based approach. It has been FDA approved for hyperammonemia and claimed to be an orphan drug. Sodium phenylbutyrate (PB) was used as an example to access the potential of carglumic acid. The anticancer activity of PB can be accounted to urea cycle activation. A study demonstrated the induction of apoptosis and in turn inhibition of proliferation by carglumic acid in various cancer viz. triple-negative breast cancer, human pancreatic cancer, lung cancer and hepatoma. Significantly, Carglumic acid promotes cell apoptosis by activating caspase 3, hence supressing tumor growth [61]. Moreover, Simvastatin (Zocor) which is an essential agent to elevate HDL (good cholesterol) and alternately, abate LDL and triglycerides (bad cholesterol) in blood, accompanied with a proper diet, has been recently found to aid in cancer treatment as well. A randomized A549 cell line study demonstrated that simvastatin and survivin (a member of the IAP family) expression are concomitant. The study depicted that simvastatin induced apoptosis via Akt signaling pathway which reverberated into survivin down-regulation in A594 cells, hence exhibiting anti-cancer effects [62]. The repurposed hits identified by pathway based strategy were given in Table 1 [63–65].
4.4. Targeted Mechanism-based approach
This approach introduces yet another drug-repositioning strategy which amalgamates data from signaling pathways, protein interaction and treatment omics to particularize the unknown workflow of drugs actions [60]. The case of Auranofin is debatably a well-known example of cancer drug repositioning issued with the help of targeted mechanism-based approach. It is basically a gold complex that has been advocated for the treatment of rheumatoid arthritis since the 1980s [66]. Treatment in the presence of gold revealed pruned malignancy rates in the patients with rheumatoid arthritis treated, with respect to the ones treated otherwise, providing a direct indication of feasibility of using auranofin for cancer therapy. Thus, several assays performed in NSCLC cell lines manifested the inhibition of AKT/PI3K/mTOR axis by auranofin, providing a clear indication of potent anti-cancer activity [67,68]. Nitroglycerin (NTG), is another illustration of a targeted mechanism-based repurposing approach. For a long time it has been used as a coronary vasodilator. Its application is also found in congestive heart failure, treatment for hypertension, and for the instigation of surgical hypotension. Recent literature summarises anti-tumor activities of NTG which is mainly attributed to nitric oxide production, though its manoeuvre against NSCLC remains explicable. There are instances of replication failure of the initially positive results, leaving scope for further exploration [69].
4.5. Target-based approach
This is a powerful technique which is embraced with high quality, instantaneous experimentation of drugs for a protein or a biomarker. It also involves in silico screening of drug libraries, viz. docking or ligand-based screening [60]. Taking the other side of the coin into account, the data might not be very reliable always particularly if the compounds have been found out from in silico library screening. Since they are more theoretical in nature, it might offer incompatible results at times. Thus, results offers an opportunity for further exploration. Clobetasol propionate (CP), a medicament that addresses dermatitis, eczema, psoriasis and a variety of skin issues, is one of the best example of repurposed candidate for NSCLC resulted from Target-based approach. A study demonstrated that, screened CP as the most potent NRF2 inhibitor among 4000 clinical compounds. Mechanistically, CP is found to play through glycogen synthase kinase 3 (GSK3) signalling, by promoting β-TrCP-dependent degradation of NRF2 and preventing nuclear accumulation. The laboratory assays unveiled that CP, either individually or in association with rapamycin (mTOR inhibitor) ceased lung tumor proliferation harbouring both KEAP1 and LKB1 mutation, which were observed in NSCLC as well [70–72].
In another study, Lam et al [73] aimed to identify the novel combination therapies for navitoclax to improve the efficacy against NSCLC by screening 640 FDA-approved drugs. The study demonstrated that benzidimidazole antihelminthic group of compounds viz. oxibendazole, albendazole, oxfendazole and mebendazole were reported to facilitate the navitoclax activity in multiple NSCLC cell lines. The data depicted that benzidimidazoles would be reliable options to homogenize with navitoclax in order to provide new combination therapies with novel mechanisms of action. Recently, ligand and energy based pharmacophore approach was employed to identify the repurposed candidates for the treatment of ALK affirmative NSCLC. The study summarized that few experimental compounds alongside FDA approved molecule, nebivolol could be repurposed for the management of NSCLC drug resistance. The study also highlights that all these compounds are able to bind effectively with ALK protein through key hydrogen bonding interaction exhibited by Met1199 [74]. Recently, cabozantinib was also used in the treatment of NSCLC particularly patients with rare oncogenic alterations, such as NTRK1 and ROS1 rearrangements [75].
4.6. Systematic in Silico Drug (Re)purposing
Improvements in computational methods and omics technology have provided new opportunity of computational drug repurposing. Valuable data, such as protein-protein interaction networks, drug-target interactions, and drug side-effect have been collected rapidly and released to the public. Although systematic computational approaches has not been yet applied to lung cancer, they will become powerful tools. We briefly overview such approaches in this section.
The most basic approach of computational drug repurposing is structure-based drug interaction prediction to new targets. Zhao et al [76] found potential inhibitors of Ebola virus by protein-ligand docking which targeted viral protein 24 (VP24) and methyltransferase (MTase). FDA-approved 2005 drugs from DrugBank [77] were docked to the predicted binding sites of the proteins using AutoDock4 [78], AutoDock Vina [79], PLANTS [80], and Surflex [81]. From the consensus of docking results, Sinefungin and Indinavir were predicted as potential inhibitors of Ebola virus infection.
Drug-target interaction (DTI) networks is an important resource for understating patterns of interacting compounds and targets, which would lead to identification of new potential targets for a drug. The assumption of a DTI network-based approach is that similar drugs are likely share targets and vice versa. It was discussed by Gao and Skolnick [82] that different proteins share similar ligand-binding pockets and pockets can be classified into a finite number of classes, which implies that a ligand can bind to multiple proteins in general [82]. Nidhi et al. [83] used extended-connectivity fingerprints (ECFP) [84] to represent compounds and trained a multiple-category naïve Bayesian model on the WOMBAT database [85] of compounds and their targets to capture features of interacting compounds and targets. When tested on the different database, MDDR [Elsevier MDL Home Page. http://www.mdli.com], the program showed a recall of 77% on known interactions and further predicted a new histone deacetylase inhibitor, [N-(2-amino-phenyl)-4-(3-hydroxypropanamido) benzamide, a close analog of known histone deacetylase inhibitor. Cao et al [86] used the PDSP Ki database [87], which composed of 514 target proteins and 3393 drug-like ligands to train random forest, a popular machine learning algorithm. Using the trained random forest, they could make 775 new connections between 67 targets and 517 drugs, among which 63% were validated from public biological resources.
In addition to DTI networks, other data such as side-effects, biological pathways, sequences and structures of target proteins, can be integrated to capture various aspects and similarities of interacting targets and drugs. Sawada et al. [88] integrated three different data types, compound features (ECFP, Chemistry Development Kit Fingerprints, and KEGG Chemical FunKCF-S), target proteins, phenotypic effects (from the FDA Adverse Event Reporting System), and disease association (the International Classification of Diseases and KEGG Disease database and KEGG Drug database).
Recently, deep-learning has also been explored to predict a new drug-target interaction. Aliper et al. [89] trained deep neural networks (DNN) on gene transcription data to predict a novel target disease of a drug. The DNN was able to re-classify drugs that were misclassified in a public database. Thus, the DNN model could serve as a drug repositioning tool. Zeng et al. [90] integrated 10 networks, namely, drug-target, drug-disease, drug-side-effect, and 7 drug-drug networks and learned interaction patterns using a deep autoencoder, which is a recent deep learning-based technique. Using the autoencoder, a new target disease for a query target drug can be predicted.
It is worth mentioning that few studies related to mutation specific inhibitors reported in the literature were also notable examples for NSCLC drug screening [91–93]. For instance, the structural and functional impact of missense mutation in EGFR, a potential drug target in cancer therapy were reported [91] in the recent years. Further, computational analysis successfully implemented in the screening of mutation specific inhibitors against EGFR and inhibitors for Bcl-2 [92, 93].
Computational approaches for drug repurposing has been rapidly developed over past years due to the improvement of machine learning methods and the increased numbers and variations of available databases. Although there are still not many drugs that were developed mainly by computational approaches, then were approved and released in market, there are already an increasing numbers of literature that report potential drugs and drug-target interactions identified by computational methods. Therefore the impact of computational approaches would only increase in coming years.
5. Combinatorial and other approaches
While no single computational biology technique is sufficient to evaluate the entire biologic range of gene–disease and drug–target relations, pioneers in this field are beginning to combine techniques to further advance computational biology. Sertraline, Bezafibrate, Mepacrine, Bosutinib and Itraconazole are the notable examples of repurposed hits emerged from combinatorial approaches [94–99]. The mechanism of action alongside the different approaches used were shown in Table 1. Adding to these approaches, few of the successful discoveries of repurposed candidate are resulted from serendipitous (or) blinded search methods in the recent years (Table 1) [100–105]. Overall, this review highlights that drug repurposing could be achieved by multiple approaches at many phases of drug development. Indeed, proper integration of different approaches able to quickly and comprehensively predict drug action with great precision.
6. Conclusions
The perennial obstacle to drug repurposing is to redefine the key drivers for success and the lack of a comprehensive library of experimental and clinical compounds suitable for testing. Perhaps, every single data, inclusive of the negative ones, should be deposited into public database. The persistent refinement of information reinforce repurposing efforts in progress and consecutively enhance the plausibility to find an efficient and skilful drug. Although repositioned hits are optimistic options with respect to NSCLC, transformation into clinical practice require contribution from multidisciplinary collaboration between oncologist and computational biologists. An extraordinary example is the Michigan Oncology Sequencing Center, which supports physicians and researchers for the integration of genomic and transcriptomic data in order to provide advanced interface for the clinical practice of cancer medicine. Further, consolidating novel and exciting tools through AI overcome these challenges and unbolt new doors for both basic cancer research and drug repurposing. Though bioinformatics-based drug repositioning is still dawning, hitherto, scientists have discerned clinically remarkable utilities for pre-existing medicaments. Together, we have taken a foundational step forward and illustrated repurposing possibilities for NSCLC by excavating recently published scientific literature from both research and clinical studies that would shed light on alternative applications for approved or failed drugs. Overall, an enlargement in drug repurposing research among the academician groups and pharmaceutical companies would certainly ramify efficient and novel anticancer therapies to regulatory market approval in the near future.
Acknowledgements
The authors (KR & VS) are grateful to Department of Science and Technology - Science and Engineering Research Board (DST-SERB) for funding the research project (File No. EMR/2016/001675). KR thank ICMR for their support by the International Fellowship for Young Biomedical Scientists Award. The authors (KR, VS & SM) thank the management of VIT, Vellore for providing the facilities to carry out this work. D.K. and W.-H. S. were partly supported by Purdue Institute of Drug Discovery, National Institutes of Health (R01GM123055), and the National Science Foundation (DMS1614777, CMMI1825941).
Conflict of interest
A conflicting interest exists when professional judgment concerning a primary interest (such as patient’s welfare or the validity of research) may be influenced by a secondary interest (such as financial gain or personal rivalry). It may arise for the authors when they have financial interest that may influence their interpretation of their results or those of others. Examples of potential conflicts of interest include employment, consultancies, stock ownership, honoraria, paid expert testimony, patent applications/registrations, and grants or other funding.
Funding source
All sources of funding should also be acknowledged and you should declare any involvement of study sponsors in the study design; collection, analysis and interpretation of data; the writing of the manuscript; the decision to submit the manuscript for publication. If the study sponsors had no such involvement, this should be stated.
References
- [1].Larsen JE, Minna JD, Molecular biology of lung cancer: clinical implications, Clin. Chest Med 32 (2011) 703–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cooper WA, Lam DC, O’Toole SA, Minna JD, Molecular biology of lung cancer, J. Thorac. Dis 5 (2013) S479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lemmon MA, Schlessinger J, Cell signaling by receptor tyrosine kinases, Cell 141 (2010) 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tidyman WE, Rauen KA, The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation, Cur Opin Genet Dev 19 (2009) 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hussain MRM, Baig M, Mohamoud HSA, Ulhaq Z, Hoessli DC, Khogeer GS, et al. , BRAF gene: from human cancers to developmental syndromes, Saudi J. Biol. Sci. 22 (2015) 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hussain MA, Ansari SA, Alqahtani MH, Shay JW, Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies, Genome Med. 8 (2016) 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gascard P, Tlsty TD, Carcinoma-associated fibroblasts: orchestrating the composition of malignancy, Genes Dev. 30 (2016) 1002–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P, Lung cancer: biology and treatment options, Biochim Biophys Acta (BBA)-Reviews on Cancer 1856. 189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zappa C, Mousa SA, Non-small cell lung cancer: current treatment and future advances, Transl. Lung Cancer Res. 5 (2016) 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Qin A, Coffey DG, Warren EH, Ramnath N, Mechanisms of immune evasion and current status of checkpoint inhibitors in non-small cell lung cancer, Cancer Med. 5 (2016) 2567–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Khalil DN, Smith EL, Brentjens RJ, Wolchok JD, The future of cancer treatment: immunomodulation, CARs and combination immunotherapy, Nat. Rev. Clin. Oncol. 13 (2016) 273–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rezvani K, Brody JD, Kohrt HE, Logan AC, Advani R, Czerwinski DK, et al. , Cancer vaccines and T cell therapy, Biol. Blood Marrow Transplant. 19 (2013) S97–S101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kazmi B, Inglefield CJ, Lewis MP, Autologous cell therapy: current treatments and future prospects, Wounds 21 (2009) 234–242. [PubMed] [Google Scholar]
- [14].Massarelli E, Papadimitrakopoulou V, Welsh J, Tang C, Tsao AS, Immunotherapy in lung cancer, Transl. Lung Cancer Res 3 (2014) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Raffoul JJ, Kucuk O, Sarkar FH, Hillman GG, Dietary agents in cancer chemoprevention and treatment, J. Oncol 2012 (2012), 749310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Donaldson MS, Nutrition and cancer: a review of the evidence for an anti-cancer diet, Nutr. J 3 (2004) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Khan N, Mukhtar H, Dietary agents for prevention and treatment of lung cancer, Cancer Lett. 359 (2015) 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rotow J, Bivona TG, Understanding and targeting resistance mechanisms in NSCLC, Nat. Rev. Cancer 17 (2017) 637. [DOI] [PubMed] [Google Scholar]
- [19].Rothschild SI, Targeted therapies in non-small cell lung cancer-beyond EGFR and ALK, Cancers 7 (2015) 930–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Pathak A, Rajappa S, Gore A, Oncogenic drivers in nonsmall cell lung cancer and resistance to epidermal growth factor receptor tyrosine kinase inhibitors, Indian J. Cancer 54 (2017) 1. [DOI] [PubMed] [Google Scholar]
- [21].Takeda M, Nakagawa K, First- and Second-Generation EGFR-TKIs Are All Replaced to Osimertinib in Chemo-Naive EGFR Mutation-Positive Non-Small Cell Lung Cancer? Int. J. Mol. Sci. 20 (2019) 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ali R, Arshad J, Palacio S, Mudad R, Brigatinib for ALK-positive metastatic non-small-cell lung cancer: design, development and place in therapy, Drug Des. Devel. Ther 13 (2019) 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roskoski R Jr, ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers, Pharmacol. Res. 121 (2017) 202–212. [DOI] [PubMed] [Google Scholar]
- [24].Ascierto PA, Kirkwood JM, Grob JJ, Simeone E, Grimaldi AM, Maio M, Mozzillo N, The role of BRAF V600 mutation in melanoma, J. Transl. Med 10 (2012) 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang Y, Sun C, Xiao G, Shan H, Tang L, Yi Y, et al. , S-nitrosylation of the Peroxiredoxin-2 promotes S-nitrosoglutathione-mediated lung cancer cells apoptosis via AMPK-SIRT1 pathway, Cell Death Dis. 10 (2019) 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim YB, Ahn YH, Jung JH, Lee YJ, Lee JH, Kang JL, Programming of macrophages by UV-irradiated apoptotic cancer cells inhibits cancer progression and lung metastasis, Cell. Mol. Immunol. (2019) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Huang TT, Lan YW, Chen CM, Ko YF, Ojcius DM, Martel J, et al. , Antrodia cinnamomea induces anti-tumor activity by inhibiting the STAT3 signaling pathway in lung cancer cells, Sci. Rep. 9 (2019) 5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kuang Y, Guo W, Ling J, Xu D, Liao Y, Zhao H, et al. , Iron-dependent CDK1 activity promotes lung carcinogenesis via activation of the GP130/STAT3 signaling pathway, Cell Death Dis. 10 (2019) 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang W, Jin G, Cai F, Chen X, Cao N, Zhang X, et al. , Extracellular signal- regulated kinase 5 increases radioresistance of lung cancer cells by enhancing the DNA damage response, Exp. Mol. Med. 51 (2019) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu DM, Zhang T, Liu YB, Deng SH, Han R, Liu T, et al. , The PAX6-ZEB2 axis promotes metastasis and cisplatin resistance in non-small cell lung cancer through PI3K/AKT signaling, Cell Death Dis. 10 (2019) 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ahmed I, Ferro A, Cohler A, Langenfeld J, Surakanti SG, Aisner J, et al. , Impact of metformin use on survival in locally-advanced, inoperable non-small cell lung cancer treated with definitive chemoradiation, J. Thorac. Dis 7 (2015) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xu H, Aldrich MC, Chen Q, Liu H, Peterson NB, Dai Q, et al. , Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality, J. Am. Med. Inform. Assoc 22 (2014) 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Amin S, Lux A, O’callaghan F, The journey of metformin from glycaemic control to mTOR inhibition and the suppression of tumour growth, Br. J. Clin. Pharmacol. 85 (2019) 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lin CC, Yeh HH, Huang WL, Yan JJ, Lai WW, Su WP, et al. , Metformin enhances cisplatin cytotoxicity by suppressing signal transducer and activator of transcription–3 activity independently of the liver kinase B1–AMP-Activated protein kinase pathway, Am. J. Respir. Cell Mol. Biol. 49 (2013) 241–250. [DOI] [PubMed] [Google Scholar]
- [35].Lin JJ, Gallagher EJ, Sigel K, Mhango G, Galsky MD, Smith CB, et al. , Survival of patients with stage IV lung cancer with diabetes treated with metformin, Am. J. Respir. Crit. Care Med. 191 (2015) 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lai SW, Liao KF, Chen PC, Tsai PY, Hsieh DPH, Chen CC, Antidiabetes drugs correlate with decreased risk of lung cancer: a population-based observation in Taiwan, Clin. Lung Cancer 13 (2012) 143–148. [DOI] [PubMed] [Google Scholar]
- [37].Chen H, Yao W, Chu Q, Han R, Wang Y, Sun J, et al. , Synergistic effects of metformin in combination with EGFR-TKI in the treatment of patients with advanced non-small cell lung cancer and type 2 diabetes, Cancer Lett. 369 (2015) 97–102. [DOI] [PubMed] [Google Scholar]
- [38].Verbaanderd C, Maes H, Schaaf MB, Sukhatme VP, Pantziarka P, Sukhatme V, et al. , Repurposing Drugs in Oncology (ReDO)—chloroquine and hydroxychloroquine as anti-cancer agents, ecancermedicalscience 11 (2017) 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rebecca VW, Amaravadi RK, Emerging strategies to effectively target autophagy in cancer, Oncogene 35 (2016) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li Y, Cao F, Li M, Li P, Yu Y, Xiang L, et al. , Hydroxychloroquine induced lung cancer suppression by enhancing chemo-sensitization and promoting the transition of M2-TAMs to M1-like macrophages, J. Exp. Clin. Cancer Res. 37 (2018) 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Millward MJ, Cantwell BMJ, Munro NC, Robinson A, Corris PA, Harris AL, Oral verapamil with chemotherapy for advanced non-small cell lung cancer: a randomised study, Br. J. Cancer 67 (1993) 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Van Nuffel AM, Sukhatme V, Pantziarka P, Meheus L, Sukhatme VP, Bouche G, Repurposing Drugs in Oncology (ReDO)—clarithromycin as an anti-cancer agent, Ecancermedicalscience 9 (2015) 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jiang J, Geng G, Yu X, Liu H, Gao J, An H, et al. , Repurposing the anti- malarial drug dihydroartemisinin suppresses metastasis of non-small-cell lung cancer via inhibiting NF-κB/GLUT1 axis, Oncotarget 7 (2016) 87271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mediavilla-Varela M, Boateng K, Noyes D, Antonia SJ, The anti-fibrotic agent pirfenidone synergizes with cisplatin in killing tumor cells and cancer-associated fibroblasts, BMC Cancer 16 (2016) 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Duan L, Shen H, Zhao G, Yang R, Cai X, Zhang L, et al. , Inhibitory effect of Disulfiram/copper complex on non-small cell lung cancer cells, Biochem. Biophys. Res. Commun. 446 (2014) 1010–1016. [DOI] [PubMed] [Google Scholar]
- [46].Tavallai M, Booth L, Roberts JL, Poklepovic A, Dent P, Rationally repurposing ruxolitinib (Jakafi®) as a solid tumor therapeutic, Front. Oncol. 6 (2016) 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang ZD, Wei SQ, Wang QY, Targeting oncogenic KRAS in non-small cell lung cancer cells by phenformin inhibits growth and angiogenesis, Am. J. Cancer Res 5 (2015) 3339. [PMC free article] [PubMed] [Google Scholar]
- [48].Rengan R, Mick R, Pryma D, Rosen MA, Lin LL, Maity AM, et al. , A phase I trial of the HIV protease inhibitor nelfinavir with concurrent chemoradiotherapy for unresectable stage IIIA/IIIB non-small cell lung cancer: a report of toxicities and clinical response, J. Thorac. Oncol 7 (2012) 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Haura EB, Rix U, Deploying ibrutinib to lung cancer: another step in the quest towards drug repurposing, J. Natl. Cancer Inst. 106 (9) (2014) dju250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Gao W, Wang M, Wang L, Lu H, Wu S, Dai B, et al. , Selective antitumor activity of ibrutinib in EGFR-mutant non–small cell lung cancer cells, J. Natl. Cancer Inst. 106 (9) (2014) dju204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jia X, Gu Z, Chen W, Jiao J, Tigecycline targets nonsmall cell lung cancer through inhibition of mitochondrial function, Fundam. Clin. Pharmacol 30 297–306. [DOI] [PubMed] [Google Scholar]
- [52].Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez- Outschoorn UE, et al. , Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease, Oncotarget 6 (2015) 4569–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Qu XA, Rajpal DK, Applications of Connectivity Map in drug discovery and development, Drug Discov. Today 17 (2012) 1289–1298. [DOI] [PubMed] [Google Scholar]
- [54].Zhang SD, Gant TW, A simple and robust method for connecting small- molecule drugs using gene-expression signatures, BMC Bioinformatics 9 (2008) 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, et al. , Drug repurposing: progress, challenges and recommendations, Nat. Rev. Drug Discov. 18 (2018) 41–58. [DOI] [PubMed] [Google Scholar]
- [56].Fortney K, Griesman J, Kotlyar M, Pastrello C, Angeli M, Sound-Tsao M, et al. , Prioritizing therapeutics for lung cancer: an integrative meta-analysis of cancer gene signatures and chemogenomic data, PLoS Comput. Biol 11 (2015), e1004068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yeh CT, Wu AT, Chang PMH, Chen KY, Yang CN, Yang SC, et al. , Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell growth and overcomes drug resistance of lung cancer, Am. J. Respir. Crit. Care Med. 186 (2012) 1180–1188. [DOI] [PubMed] [Google Scholar]
- [58].Yuen T, Stachnik A, Iqbal J, Sgobba M, Gupta Y, Lu P, et al. , Bisphosphonates inactivate human EGFRs to exert antitumor actions, Proc Natl Acad Sci U S A 111 (2014) 17989–17994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Srirangam A, Milani M, Mitra R, Guo Z, Rodriguez M, Kathuria H, et al. , The human immunodeficiency virus protease inhibitor ritonavir inhibits lung cancer cells, in part, by inhibition of survivin, J. Thorac. Oncol 6 (2011) 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jin G, Wong ST, Toward better drug repositioning: prioritizing and integrating existing methods into efficient pipelines, Drug Discov. Today 19 (2014) 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen CT, Chen YC, Yamaguchi H, Hung MC, Carglumic acid promotes apoptosis and suppresses cancer cell proliferation in vitro and in vivo, Am. J. Cancer Res 5 (2015) 3560. [PMC free article] [PubMed] [Google Scholar]
- [62].Hwang KE, Na KS, Park DS, Choi KH, Kim BR, Shim H, et al. , Apoptotic induction by simvastatin in human lung cancer A549 cells via Akt signaling dependent down-regulation of survivin, Invest New Drug 29 (2011) 945–952. [DOI] [PubMed] [Google Scholar]
- [63].Saxena A, Becker D, Preeshagul I, Lee K, Katz E, Levy B, Therapeutic effects of repurposed therapies in non-small cell lung cancer: what is old is new again, Oncologist 20 (2015) 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Park IH, Kim JY, Jung JI, Han JY, Lovastatin overcomes gefitinib resistance in human non-small cell lung cancer cells with K-Ras mutations, Invest. New Drugs 28 (2010) 791–799. [DOI] [PubMed] [Google Scholar]
- [65].Huang CH, Wu MY, Chang PMH, Huang CY, Ng KL, In silico identification of potential targets and drugs for non-small cell lung cancer, IET Syst. Biol 8 (2014) 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chaffman M, Brogden RN, Heel RC, Speight TM, Avery GS, Auranofin, Drugs 27 (1984) 378–424. [DOI] [PubMed] [Google Scholar]
- [67].Li H, Hu J, Wu S, Wang L, Cao X, Zhang X, et al. , Auranofin-mediated inhibition of PI3K/AKT/mTOR axis and anticancer activity in non-small cell lung cancer cells, Oncotarget 7 (2016) 3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Roder C, Thomson MJ, Auranofin: repurposing an old drug for a golden new age, Drugs R. 15 (2015) 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sukhatme V, Bouche G, Meheus L, Sukhatme VP, Pantziarka P, Repurposing Drugs in Oncology (ReDO)—nitroglycerin as an anti-cancer agent, ecancermedicalscience 9 (2015) 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Choi EJ, Jung BJ, Lee SH, Yoo HS, Shin EA, Ko HJ, et al. , A clinical drug library screen identifies clobetasol propionate as an NRF2 inhibitor with potential therapeutic efficacy in KEAP1 mutant lung cancer, Oncogene 36 (2017) 5285. [DOI] [PubMed] [Google Scholar]
- [71].Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. , Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities, Cancer Discov. 5 (2015) 860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kaufman JM, Amann JM, Park K, Arasada RR, Li H, Shyr Y, Carbone DP, LKB1 Loss induces characteristic patterns of gene expression in human tumors associated with NRF2 activation and attenuation of PI3K-AKT, J. Thorac. Oncol 9 (2014) 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lam LT, Zhang H, Xue J, Leverson JD, Bhathena A, Antihelminthic benzimidazoles potentiate navitoclax (ABT-263) activity by inducing Noxa- dependent apoptosis in non-small cell lung cancer (NSCLC) cell lines, Cancer Cell Int. 15 (2015) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].James N, Shanthi V, Ramanathan K, Discovery of novel anaplastic lymphoma kinase inhibitors: structure and energy based pharmacophore strategy, J. Theor. Comput. Chem. 18 (3) (2019), 1950014. [Google Scholar]
- [75].Chong CR, Bahcall M, Capelletti M, Kosaka T, Ercan D, Sim T, et al. , Identification of existing drugs that effectively target NTRK1 and ROS1 rearrangements in lung cancer, Clin. Cancer Res. 23 (2017) 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhao Z, Martin C, Fan R, Bourne PE, Xie L, Drug repurposing to target Ebola virus replication and virulence using structural systems pharmacology, BMC Bioinformatics 17 (2016) 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, et al. , DrugBank: a knowledgebase for drugs, drug actions and drug targets, Nucleic Acids Res. 36 (Database issue) (2008) D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ, AutoDock4 and AutoDockTools: automated docking with selective receptor flexibility, J. Comput. Chem. 30 (2009) 2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Trott O, Olson AJ, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem. 31 (2010) 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Korb O, Stuzle T, Exner TE, Empirical scoring functions for advanced protein- ligand docking with PLANTS, J. Chem. Inf. Model. 49 (2009) 84. [DOI] [PubMed] [Google Scholar]
- [81].Jain AN, Surflex-Dock 2.1: robust performance from ligand energetic modelling, ring flexibility and knowledge-based search, J. Comput. Aided Mol. Des. 21 (2007) 281. [DOI] [PubMed] [Google Scholar]
- [82].Gao M, Skolnick J, A comprehensive survey of small-molecule binding pockets in proteins, PLoS Comput. Biol 9 (2013), e1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Nidhi GM, Davies JW, Jenkins JL, Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogeonics databases, J. Chem. Inf. Model. 46 (2006) 1124. [DOI] [PubMed] [Google Scholar]
- [84].Morgan HL, The generation of a unique machine description for chemical Structures-A technique developed at chemical abstracts service, J. Chem. Doc. 5 (1965) 107. [Google Scholar]
- [85].Olah M, Mracec M, Ostopovici L, Rad R, Bora A, Hadaruga N, et al. , WOMBAT: world of molecular bioactivity. Cheminformatics in Drug Discovery; Oprea TI, Ed., Wiley-VCH, New York, 2004, pp. 223–239. [Google Scholar]
- [86].Cao DS, Liang DZ, Deng Z, Hu QN, He M, Xu QS, et al. Genome-scale screening of drug-target associations relevant to Ki using a chemogenomics approach PLoS One; 8:e57680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK, BindingDB: a web-accesible database of experimentally determine protein-ligand binding affinities, Nucleic Acids Res. 35 (2007) D198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sawada R, Iwata H, Mizutani S, Yamanishi Y, Target-based drug repositioning using large-scale chemical-protein interactome data, J. Chem. Inf. Model. 55 (2015) 2717. [DOI] [PubMed] [Google Scholar]
- [89].Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A, Deep learning applications of drugs and drug repurposing using transcriptomic data, Mol. Pharm. 13 (2016) 2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zeng X, Zhu S, Liu X, Zhou Y, Nussinov R, Cheng F, deepDR: a network-based deep learning approach to in silico drug repositioning, Bioinformatics (2019). In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Anoosha P, Sakthivel R, Gromiha MM, Investigating mutation-specific biological activities of small molecules using quantitative structure-activity relationship for epidermal growth factor receptor in cancer, Mutat. Res. 806 (2017) 19–26. [DOI] [PubMed] [Google Scholar]
- [92].Kanakaveti V, Rathinasamy S, Rayala SK, Gromiha M, Forging new scaffolds from old: combining scaffold hopping and hierarchical virtual screening for identifying novel Bcl-2 inhibitors, Curr. Top. Med. Chem. 19 (13) (2019) 1162–1172. [DOI] [PubMed] [Google Scholar]
- [93].Kanakaveti V, Anoosha P, Sakthivel R, Rayala SK, Gromiha MM, Influence of amino acid mutations and small molecules on targeted inhibition of proteins involved in Cancer, Curr. Top. Med. Chem. 19 (6) (2019) 457–466. [DOI] [PubMed] [Google Scholar]
- [94].Jiang X, Lu W, Shen X, Wang Q, Lv J, Liu M, et al. , Repurposing sertraline sensitizes non–small cell lung cancer cells to erlotinib by inducing autophagy, JCI Insight 3 (11) (2018), e98921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Kim J, Vasu VT, Mishra R, Singleton KR, Yoo M, Leach SM, et al. , Bioinformatics-driven discovery of rational combination for overcoming EGFR-mutant lung cancer resistance to EGFR therapy, Bioinformatics 30 (2014) 2393–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Liu X, Yang X, Chen X, Zhang Y, Pan X, Wang G, et al. , Expression profiling identifies bezafibrate as potential therapeutic drug for lung adenocarcinoma, J. Cancer 6 (2015) 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Huang CH, Chang PMH, Hsu CW, Huang CYF, Ng KL, Drug repositioning for non-small cell lung cancer by using machine learning algorithms and topological graph theory, In BMC bioinformatics BioMed Central 17 (2016) S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Shim JS, Liu JO, Recent advances in drug repositioning for the discovery of new anticancer drugs, Int. J. Biol. Sci. 10 (2014) 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Aftab BT, Dobromilskaya I, Liu JO, Rudin CM, Itraconazole inhibits angiogenesis and tumor growth in non–small cell lung cancer, Cancer Res. 71 (2011) 6764–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang B, Yu W, Guo J, Jiang X, Lu W, Liu M, Pang X, The antiparasitic drug, potassium antimony tartrate, inhibits tumor angiogenesis and tumor growth in nonsmall-cell lung cancer, J. Pharmacol. Exp. Ther. 352 (2015) 129–138. [DOI] [PubMed] [Google Scholar]
- [101].Zhang L, He M, Zhang Y, Nilubol N, Shen M, Kebebew E, Quantitative high-throughput drug screening identifies novel classes of drugs with anticancer activity in thyroid cancer cells: opportunities for repurposing, J. Clin. Endocrinol. Metab. 97 (2012) E319–E328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Davies AM, Lara PN, Mack PC, Gandara DR, Incorporating bortezomib into the treatment of lung cancer, Clin. Cancer Res. 13 (2007) 4647–4651. [DOI] [PubMed] [Google Scholar]
- [103].Chanvorachote P, Pongrakhananon V, Ouabain downregulates Mcl-1 and sensitizes lung cancer cells to TRAIL-induced apoptosis, Am. J. Physiol., Cell Physiol. 304 (2012) 263–272. [DOI] [PubMed] [Google Scholar]
- [104].Li J, Huang Y, Gao Y, Wu H, Dong W, Liu L, Antibiotic drug rifabutin is effective against lung cancer cells by targeting the eIF4E-β-catenin axis, Biochem. Biophys. Res. Commun. 472 (2016) 299–305. [DOI] [PubMed] [Google Scholar]
- [105].Kim MO, Choe MH, Yoon YN, Ahn J, Yoo M, Jung KY, et al. , Antihelminthic drug niclosamide inhibits CIP2A and reactivates tumor suppressor protein phosphatase 2A in non-small cell lung cancer cells, Biochem. Pharmacol. 144 (2017) 78–89. [DOI] [PubMed] [Google Scholar]