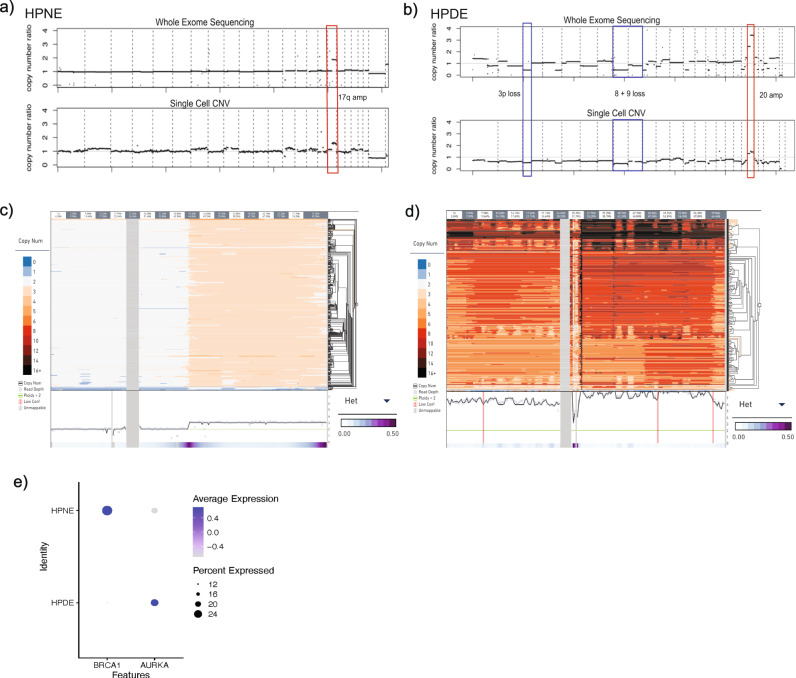
Fig. 2. Genomic characterization of HPDE and HPNE normal control cell lines.
a scCNV comparison to WES of HPNE cell line depicting amplified 17q as the only notable CNV event. b scCNV comparison to WES of HPDE cell line. CNV events representing losses of chromosomal arms 3p, 8p, and 9p and amplifications at chromosome 20. c scCNV high-resolution cell phylogeny of HPNE for all chromosome 17 locations showing ploidy = 3 for all cells at 17q arm, region inclusive of BRCA1 loci. Columns indicate chromosomal intervals measured in Mb, rows indicate individual cells. Chromosomal regions depicted (labeled along top x-axis) are representative of CNV segments outlined in red in Fig. 1d. d scCNV high-resolution cell phylogeny of HPDE for all chromosome 20 locations showing ploidy >3 (as high as 13 in some cells at some locations) for all cells. Columns indicate chromosomal intervals measured in Mb, rows indicate individual cells. Chromosomal regions depicted (labeled along top x-axis) are representative of CNV segments outlined in red in Fig. 1e. Corresponding scRNA data shows elevated expression of AURKA, located within amplified HPDE region as a potential target of amplification. e scRNAseq data of HPNE and HPDE cell lines shows elevated expression of BRCA1 in >20% of HPNE cells, increased AUKRA expression in >20% of HPDE cells.