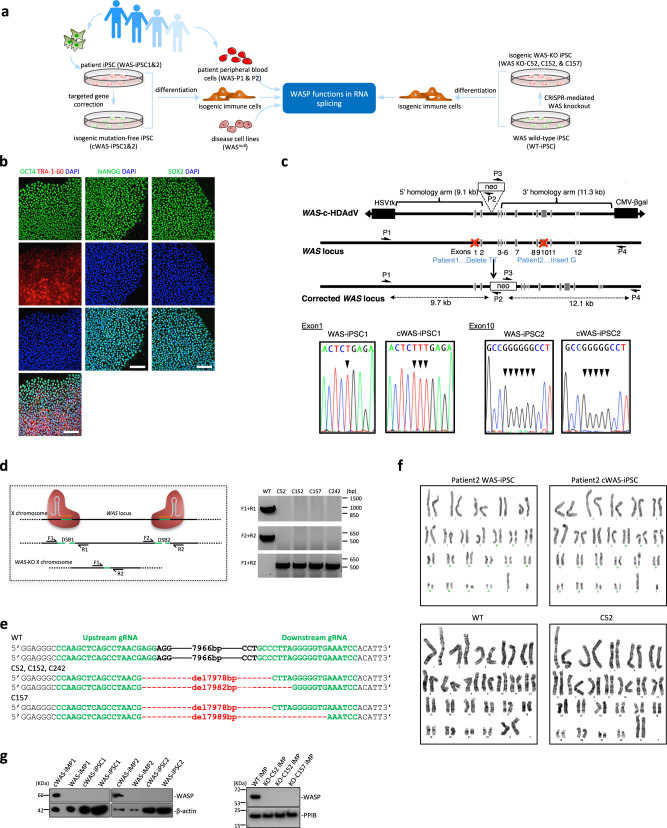
Fig. 1. Generation of isogenic iPSC models of WAS.
a Schematic diagram of the overall study design (created with smart.servier.com). b Representative immunofluorescence images of pluripotency markers OCT4, Tra-1-60, NANOG, and SOX2 in WAS-iPSC. DNA was stained with DAPI. Bar, 100 mm. The experiment was repeated 4 times independently. c Schematic molecular representation of WAS gene correction with gene-correction vector (WAS-c-HDAdV). The primers for PCR verification of gene targeting are shown as arrows (P1, P2, P3, and P4). HSVtk stands for herpes simplex virus thymidine kinase gene cassette used for negative selection; neo stands for neomycin-resistant gene cassette used for positive selection; CMV-βgal: β-gal expression cassette for determination of HDAdV titer; Red crosses mark the mutations c.107_108del in exon 1 and c.1271dupG in exon 10 present in Patient 1 and Patient 2, respectively. d The schematic of the CRISPR design, and representative PCR genotyping results of four iPSC clones with the entire WAS gene deleted. DBS: double-strand break, the experiment was repeated three times independently. bp base pair. e Sanger sequencing of the PCR product of F1 + R2 shows the CRISPR-induced deletion breakpoints in four KO iPSC clones. f Representative karyotyping analysis revealed normal karyotypes in randomly selected clones of WAS-iPSC lines, either before or after gene editing. g Western blot analysis of WASP protein in iMPs and iPSCs. Beta-actin or PPIB was used as a loading control. kDa: kilodalton (applicable to all western blot images).