Abstract
Antimicrobial peptides (AMPs) are key to defence against infection in plants and animals. Use of AMP mutations in Drosophila has now revealed that AMPs can additively or synergistically contribute to defence in vivo. However, these studies also revealed high specificity, wherein just one AMP contributes an outsized role in combatting a specific pathogen. Here, we show the Drosocin locus (CG10816) is more complex than previously described. In addition to its namesake peptide ‘Drosocin’, it encodes a second mature peptide from a precursor via furin cleavage. This peptide corresponds to the previously uncharacterized ‘Immune-induced Molecule 7’. A polymorphism (Thr52Ala) in the Drosocin precursor protein previously masked the identification of this peptide, which we name ‘Buletin’. Using mutations differently affecting Drosocin and Buletin, we show that only Drosocin contributes to Drosocin gene-mediated defence against Enterobacter cloacae. Strikingly, we observed that Buletin, but not Drosocin, contributes to the Drosocin gene-mediated defence against Providencia burhodogranariea, including an importance of the Thr52Ala polymorphism for survival. Our study reveals that the Drosocin gene encodes two prominent host defence peptides with different specificity against distinct pathogens. This finding emphasizes the complexity of the Drosophila humoral response and demonstrates how natural polymorphisms can affect host susceptibility.
Keywords: antimicrobial peptide, host–pathogen interactions, Drosophila, immunity
1. Introduction
The ability to rapidly combat pathogens is critical to organism health and survival. Organisms sense natural enemies through pattern recognition receptors, triggering the activation of core immune signalling pathways. These pathways regulate the expression of a plethora of immune effectors that provide a first line of innate defence. It was generally thought that innate immune effectors act together as a cocktail to kill microbes. However recent studies have challenged this view, revealing an unexpectedly high degree of specificity in the effector response to infection [1–3].
Chief among immune effectors are antimicrobial peptides (AMPs), host-encoded antibiotics that exhibit microbicidal activities [1,2,4,5]. Insects, and particularly the genetically tractable model Drosophila, have been especially fruitful in identifying and characterizing AMP potency and function [4,6–9]. In Drosophila, systemic infection triggers the expression of a battery of antimicrobial peptides that are secreted into the haemolymph by the fat body to transform this compartment into a potent microbicidal environment. This systemic AMP response is tightly regulated by two signalling cascades: the Toll and Imd pathways. These two pathways are similar to mammalian Toll-like receptor and tumour necrosis factor alpha/nuclear factor kappa B signalling that regulate the inflammatory response [10,11]. They are differentially activated by different classes of microbes. The Toll pathway is predominantly instigated after sensing infection by Gram-positive bacteria and fungi, while the Imd pathway is especially responsive to Gram-negative bacteria and some Gram-positive bacteria with diaminopimelic acid-type peptidoglycan [11–13]. The expression of each AMP gene is complex, receiving differential input from either pathway, with most AMPs being at least somewhat co-regulated during the systemic immune response [14–16].
In Drosophila, several families of AMPs contribute downstream of Toll and Imd. This includes the Cecropin, Attacin, Diptericin, Defensin, Metchnikowin, Drosomycin, Baramicin and Drosocin gene families [1,3,4]. Other host defence peptide families include Daisho and Bomanin, which are important for defence, but in vitro killing activity is yet to be shown [17,18]. How these immune effectors contribute individually or collectively to host defence remains poorly understood. Use of single and compound mutants has revealed that defence against some pathogens relies on the collective contributions of multiple AMP families. However, recent studies have also shown that single defence peptides can play highly specific and important roles during infection. In one case, Diptericins are the critical AMP family for surviving infection by Providencia rettgeri bacteria. This specificity is so remarkable that flies collectively lacking five other AMP gene families nevertheless resist P. rettgeri infection like the wild-type [6], while even a single amino acid change in one Diptericin gene can cause pronounced susceptibility to P. rettgeri [19]. Studies on Toll effector genes such as Bomanins, Daishos or Baramicin A have also found deletion of single gene families can cause strong susceptibilities against specific fungal species [18,20], or mediate general defences against broad pathogen types [17,21]. Lastly, loss of the gene Drosocin causes a specific and pronounced susceptibility to infection by Enterobacter cloacae [6], agreeing with Drosocin peptide activity in vitro [22]. Unlike the example with Diptericins and P. rettgeri, other AMPs also contribute collectively to defence against En. cloacae [23].
Many AMP genes encode precursor proteins with multiple peptide products processed by furin cleavage [20]. This was initially shown for the Apidaecin gene of honeybees, which produces nine Apidaecin peptides from a single precursor [24]. Drosophila also encodes many AMPs with polypeptide precursors. Examples include AMPs of the Attacin and Diptericin gene families [25,26] or Baramicin A which encodes three kinds of unique peptide products on a single precursor protein [1,20,27]. Meanwhile, the precursor protein of the nematode AMP ‘NLP29’ is cleaved into six similar glycine-rich peptides [28,29]. To our knowledge, the independent contributions of sub-peptides from a polypeptide AMP gene has so far never been addressed.
In this study, we reveal that the Drosocin gene (CG10816) encodes not only the antibacterial Drosocin peptide but also another host defence peptide produced by furin cleavage of the Drosocin precursor protein. We name this peptide Buletin, and show that it corresponds to IM7, an inducible peptide first identified in 1998 by MALDI-TOF analysis whose gene counterpart was never identified [30]. Using a new mutation affecting only the Drosocin peptide and not Buletin, we show that these two peptides contribute independently to defence against different microbes. Survival analyses show that while Drosocin specifically affects defence against En. cloacae, Buletin specifically affects defence against Providencia burhodogranariea. Moreover, a previously identified polymorphic site in Buletin (Thr52Ala described in [31]) mirrors the susceptibility effect of Buletin deletion to P. burhodogranariea. We, therefore, uncover a striking example where an AMP-encoding gene produces two peptides with distinct activities. The Drosocin gene is also an example of how an AMP polymorphism can significantly affect the host defence against a specific microbe. Alongside recent findings using Diptericin and P. rettgeri, our results highlight how AMP evolution is probably driven by differential activity against ecologically relevant microbes.
2. Results
For clarity of discussion: we will use the shorthand Drc (with a ‘c’, no italics) to refer to the mature Drosocin peptide. Whenever possible, we will use ‘Drosocin gene’ to refer to the genomic locus (common shorthand Dro, with an ‘o’, italicized).
(a) . The Drosocin gene encodes IM7
Previous proteomic analyses of haemolymph from infected Drosophila revealed several immune-induced molecules (IMs) [30]. These molecules were annotated as IM1-IM24 according to their mass, and over time each of these IMs was associated with a host defence peptide gene [17,18,20,32]. At this point, only one of the 24 original IMs remains unknown: IM7. Previous efforts were unable to link this 2307 Da peptide to a gene in the Drosophila reference genome. However, during our studies, we noticed that IM7 was absent in flies lacking 14 AMP genes, indicating that it is probably produced by one of these genes [6,23]. We repeated these MALDI-TOF proteomic experiments with haemolymph samples from flies carrying systematic combinations of AMP mutations, ultimately honing in on the gene Drosocin (Dro). Two independent Dro gene mutants (DroSK4 and Dro-AttABSK2) both lack IM7 in MALDI-TOF peptidomic analysis (figure 1).
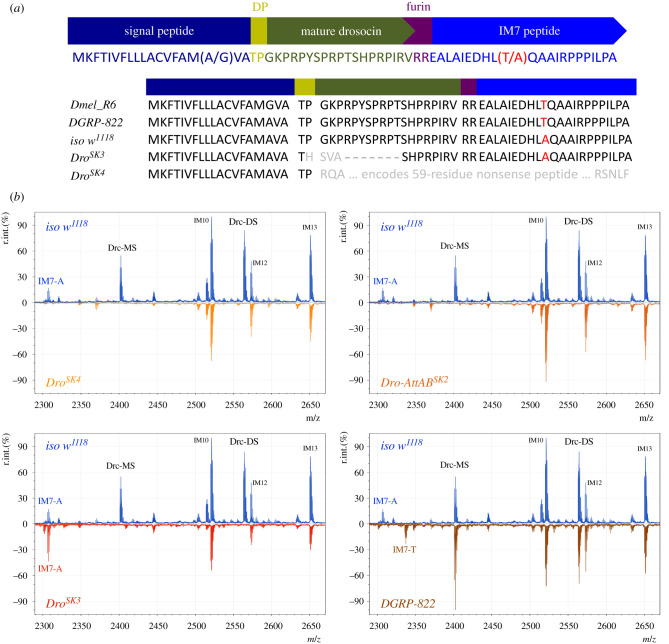
Figure 1.
The Dro gene encodes a polypeptide including both Drc and IM7. (a) Overview of the precursor protein structure of the Dro gene. The Thr52Ala polymorphism in IM7 was noted previously [31]. Here we include an alignment of the Drosocin precursor protein between the Dmel_R6 reference genome and sequences from iso w1118, DroSK3, DroSK4 and DGRP-822 flies. "DP" = dipeptidyl peptidase cleavage motif. "furin" = furin cleavage site motif. (b) MALDI-TOF proteomic data from immune-challenged flies shows that both Drc (Drc-MS, Drc-DS) and the 2307 Da peak of IM7 is absent in DroSK4 and Dro-AttABSK2 flies. The frameshift present in DroSK3 removes the Drc peptide, but does not prevent the secretion of IM7. Threonine-encoding IM7 appears in DGRP-822 (2337 Da), alongside loss of the 2307 Da peak. (Online version in colour.)
The Dro gene was initially identified as a single open reading frame gene encoding the Drc peptide. Drc is an O-glycosylated Proline-rich peptide that binds bacterial DnaK/Hsp70 similar to other Proline-rich insect AMPs [22,33–36]. Mature Drc requires O-glycosylation for activity, which involves the biochemical linking of either mono- (MS), di- (DS), or rarely tri-saccharide (TS) groups to the Threonine at position 11 of the Drc peptide [22,32]. These different O-glycosylations yield peptides with different mature masses of 2401, 2564, and 2767 Da (Drc-MS, -DS and -TS, respectively). Unmodified Drc peptide has an expected mass of 2199 Da, which is not an intuitive match for the 2307 Da peak of IM7, even considering other post-translational modifications. This suggests that another element of the Dro gene encodes IM7.
(b) . IM7 is the C-terminus product of the Drosocin precursor protein
It is puzzling that IM7 could not be annotated to the Dro gene given that the nucleotide sequence has been known for decades. One previous study noted that the Dro gene was probably cleaved at a furin-like cleavage site, and had a small undescribed C-terminal peptide [25]. Lazzaro & Clark [31] further described a polymorphism in the Dro gene encoding either a Threonine or Alanine at residue 52 in the C-terminus of the precursor protein sequence (Thr52Ala). The Drosophila melanogaster reference genome encodes the Threonine version of this polymorphism. Using the sequence of the reference genome, the Drosocin precursor C-terminus mature mass would be 2337 Da without considering post-translational modifications. If we instead substitute an Alanine at this site, the predicted mass of the Drosocin precursor C-terminus becomes 2307 Da, exactly matching the observed mass of IM7. We confirmed that our wild-type DrosDel isogenic genetic background encodes an Alanine allele both by Sanger sequencing and liquid chromatography-mass spectrometry proteomics. We next performed MALDI-TOF on the haemolymph of flies from DGRP strain 822 (DGRP-822), which encodes a Threonine in its C-terminus. Exactly matching prediction, DGRP-822 flies lack the 2307 Da IM7 peak, and instead have a 2337 Da peak that appears after infection (figure 1b).
Serendipitously, while generating Dro gene mutants using CRISPR-Cas9 we recovered a complex aberrant locus (DroSK3) that disrupts 11 amino acid residues of the mature Drc peptide, including its critical O-glycosylated Threonine (figure 1a). However the DroSK3 deletion later continues in the same reading frame, including the RVRR furin cleavage site and C-terminus. Thus we suspected that the C-terminal peptide would be secreted normally in DroSK3 flies. When we ran MALDI-TOF analysis on immune-induced haemolymph from DroSK3 flies, we recovered a signal that all but confirmed the identity of the Dro gene C-terminus: DroSK3 flies lacked the Drc-MS and Drc-DS peaks, but the 2307 Da peak corresponding to IM7 remained immune-inducible (figure 1b).
Taken together, we reveal that the Dro gene encodes two peptides: Drc and IM7, which are produced from a precursor protein by cleavage at a canonical furin cleavage site. IM7 is a 22-residue peptide with a net anionic charge (−1.9 at pH = 7) that does not share overt similarity with Drc (+5.1 at pH = 7), though both peptides are Proline-rich. A naturally occurring polymorphism previously obscured the annotation of IM7 as a Dro gene product. This analysis was greatly facilitated by the use of newly available AMP mutations. We name this C-terminal peptide Buletin (Btn) after Philippe Bulet, whose dedicated efforts in the 1980s–1990s characterized many of the Drosophila AMPs including Drosocin [4,22,37].
(c) . Drosocin, but not Buletin, is responsible for the Drosocin gene-mediated defence against Enterobacter cloacae
Previous studies have suggested that flies lacking just the Dro gene can resist infection by most bacteria, but are specifically susceptible to infection by En. cloacae [6], and also somewhat Escherichia coli [38] and P. burhodogranariea [6]. The fact that the Dro gene encodes not one but two peptides raises the question of the specific contribution of these two peptides to previously observed Dro gene effects. Therefore, we took advantage of the DroSK3 and DroSK4 mutations that differently affect the Drc and Btn peptides (figure 1a) to explore the respective role(s) these peptides play by comparing the survival of these mutants to different infections. We focused our screen on a panel of Gram-negative bacteria of interest: En. cloacae β12 bacteria that Dro gene mutants are specifically susceptible to [6,23], a recently-isolated Acetobacter sp. that can kill AMP mutant flies [39], E. coli 1106 suggested to be affected by the Dro gene [22,38], and P. burhodogranariea strain B where the Dro gene was shown to contribute to defence alongside other AMPs [6]. All experiments were performed with wild-type and mutant flies that were isogenized in the DrosDel genetic background according to Ferreira et al. [40].
We found that individual Dro gene mutants (both DroSK3 and DroSK4) were not overtly susceptible to infection by E. coli 1106 or Acetobacter sp. ML04.1 (electronic supplementary material, figure S1). We could also repeat our previous findings that DroSK4 and Dro-AttABSK2 flies were highly susceptible to En. cloacae infection, causing 40–50% mortality by 3 days after infection. Importantly, use of DroSK3 flies that lack Drc but produce Btn confirms that this susceptibility is principally caused by a loss of Drc peptide and not Btn (figure 2a): DroSK4 and Dro-AttABSK2 flies lacking both Drc and Btn were only slightly more susceptible than DroSK3 flies lacking Drc alone, a difference that was not statistically significant (DroSK4 and Dro-AttABSK2 comparisons to DroSK3, p > 0.05 in both cases).
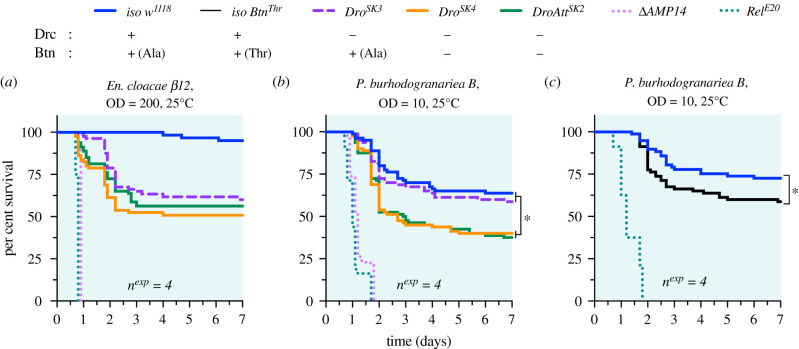
Figure 2.
Mutations affecting Buletin cause a specific susceptibility to P. burhodogranaria. (a) DroSK3 flies succumb to infection by En. cloacae slightly later than either DroSK4 or Dro-AttABSK2 flies that lack both Drc and Btn. The ultimate rate of mortality is comparable (p > 0.05 in comparison between these various Dro mutants). (b) Drosocin mutants that retain Btn (DroSK3) survive infection by P. burhodogranariea better than flies lacking both Drc and Btn (DroSK4, DroAttSK2). (c) Wild-type flies with the Threonine allele of the Btn Thr52Ala polymorphism phenocopy the effect of Btn deletion compared to Alanine-encoding iso w1118 in defence against P. burhodogranariea. OD, optical density. (Online version in colour.)
Thus, comparison of mutants lacking Drc, or both Drc and Btn, reveals that the Dro gene-mediated defence against En. cloacae is specifically mediated by the Drc peptide. Meanwhile, Btn does not seem to contribute to defence against this bacterial infection in a significant way.
(d) . Buletin, but not Drosocin, is important for survival to Providencia burhodogranariea infection
We previously found that the Dro gene could contribute to defence against P. burhodogranariea synergistically alongside Diptericins and Attacins [6]. We next assessed the contribution of our different Dro gene mutants to defence against P. burhodogranariea. To our surprise, the presence or absence of Btn causes a pronounced survival difference after infection by P. burhodogranariea: DroSK3 flies that still produce Btn survive as wild-type, while DroSK4 or Dro-AttABSK2 flies suffer significantly increased mortality (figure 2b). This trend is the opposite of what is observed after infection with En. cloacae: Drc does not play an important role in defence against P. burhodogranariea, but Btn does. As emphasized by the greater susceptibility of AMP-deficient ΔAMP14 and Imd-deficient RelE20 flies (figure 2b), Btn deficiency explains only part of the susceptibility to P. burhodogranariea. This is consistent with our previous study, which showed that the Dro gene contributes to defence against this bacterium alongside the contributions of Diptericin and Attacin genes.
Collectively, our study shows that the Dro locus encodes two host-defence peptides with distinct activities in vivo. This reinforces the notion that innate immune effectors can have very specific roles in vivo.
(e) . The Thr52Ala polymorphism affects Buletin activity against Providencia burhodogranariea in vivo
The existence of a Threonine/Alanine polymorphic residue in Btn in natural fly populations suggests an arms race between Btn and naturally occurring pathogens. Such polymorphisms are common in AMP genes, and are proposed to reflect host-pathogen coevolutionary selection [41,42]. The P. burhodogranariea strain used in this study was originally isolated from the haemolymph of wild-caught flies [43], suggesting it is an ecologically relevant microbe to D. melanogaster. This prompted us to investigate the contribution of this polymorphism in defence against P. burhodogranariea. We next isolated a Btn-Threonine allele (BtnThr) that we introgressed into the DrosDel background over seven generations. We infected isogenic BtnThr and BtnAla (i.e. iso w1118) flies with P. burhodogranariea to determine if the Btn polymorphism impacts survival. In these experiments, iso BtnThr flies suffered an approximately 15% increase in mortality compared to iso w1118 flies with BtnAla (figure 2c, p = 0.037). The Cox survival hazard ratio is a measure of effect size. The hazard ratio of DroSK4 versus DroSK3 flies (figure 2b) and iso BtnThr versus iso w1118 (figure 2c) is nearly-identical (hazard ratios: DroSK4-DroSK3 = 0.590, BtnThr-iso w1118: = 0.584). Thus the effect size of changing the Btn allele from Alanine to Threonine causes the same hazard ratio difference as the effect of Btn deletion.
We, therefore, uncover an important role of Btn in defence against P. burhodogranariea, and reveal that the Btn Thr52Ala polymorphism impacts survival against this ecologically relevant pathogen. Alongside the effect of a polymorphism in Diptericin on survival to P. rettgeri [19], here we provide a second example of how a polymorphic residue in an AMP gene significantly impacts survival.
3. Discussion
Here we show that the Dro gene encodes two peptides with distinct activities in vivo. Buletin was not annotated previously owing to a polymorphism that masked the identity of this second peptide. Most immune studies have used Drosophila strains that encode the BtnAla allele (e.g. Oregon-R [30], w1118 [44], DrosDel [6] or Canton-S [25]), while the D. melanogaster reference genome encodes the BtnThr allele. The Dro gene produces a precursor protein cleaved in two locations: (i) after the signal peptide at a two-residue dipeptidyl peptidase site that is nibbled off of the N-terminus of mature Drc (electronic supplementary material, figure S3, similar sites noted in [20,45]); and (ii) at a furin cleavage motif that separates the Drc and Btn peptides (‘RVRR’ in the Drosocin precursor protein). Both cleavage motifs are common in AMPs, including Drosophila Attacins, Defensins, Diptericins and Baramicins, which all encode mature peptides separated by furin cleavage sites [1,20,25].
The Dro gene is restricted to the genus Drosophila [46]. However phylogenetic inference for AMPs is difficult owing to their short size [3,47], and functional analogues of the Drc peptide that may share an evolutionary history are described in many holometabolous insects [36,48]. It is therefore noteworthy that the range of Buletin is far more restricted: Buletin-like peptides are found only in Dro genes of fruit flies ranging from the Melanogaster to Obscura groups, and not in outgroup Drosophila species (electronic supplementary material, figure S2). The Buletin peptide is therefore an evolutionary novelty of the Dro gene C-terminus. The Thr52Ala polymorphism in Buletin is probably maintained by balancing selection [42], similar to a specific susceptibility for the Arginine variant of a Serine/Arginine polymorphism in Diptericin for defence against P. rettgeri [19]. The reason behind these polymorphisms is unclear but could rely on trade-offs in immune defence and other functions [2,49]. Trade-offs have been especially well characterized in the fly cellular immune response where higher haemocyte numbers improve host resistance to parasitoid wasps but reduce larval competitive ability [50,51]. This is probably owing to the high metabolic cost imposed by higher haemocyte numbers, reducing available fat body lipid stores [52]. Another example is the trade-off between reproduction and immunity, as both sides impose a high metabolic demand on the insect fat body in females [53–55]. While there is renewed attention on how positive selection promotes AMP polymorphisms, we know less about the evolutionary forces that maintains these alternate alleles [41,56,57]. A simple interpretation for why AMP polymorphisms exist might be that alternate residues improve resistance against specific pathogens, resulting in the maintenance of two alleles with different pathogen-specific competences. This is tempting to speculate given the Providencia species used here and in earlier Drosophila studies were isolated from wild flies [43]. Likewise, En. cloacae has sometimes been recovered in the microbiome of D. melanogaster [58]. However these proposals lack conclusive proof, as the precise logic driving the DptA or Btn polymorphisms is currently defined only by one residue being better for defence against one specific Providencia bacteria. Currently, we have no evidence for an alternate allele to promote defence against another pathogen. Moreover, we cannot exclude that these polymorphisms could relate to AMP roles beyond infection, as recent studies have found surprising roles for AMPs in things like memory formation and behavioural regulation [28,59–62]. For now, the evolutionary purpose of the DptA and Btn alternate alleles remains unknown.
The Drc and Btn peptides are not homologous, although both are rich in Proline residues. However, Drosocin is O-glycosylated and has a strong cationic charge (+5.1 at pH = 7), while Buletin is unmodified and has a net anionic charge (−1.9 at pH = 7). AlphaFold predicts Buletin to have an α-helical structure [63]. We screened for Buletin activity in vitro diluted in Luria Broth (LB) according to Wiegand et al. [64]. However, in our conditions, we found no effect of Buletin using either BtnThr or BtnAla against P. burhodogranariea or E. coli, even when co-incubated with sub-lethal concentrations of Cecropin (Sigma) (electronic supplementary material, figure S4). It is possible that Buletin contributes to host defence alongside a cofactor, or protects the host from a virulence factor secreted by P. burhodogranariea. It may even be that Buletin is required for some role in physiology unrelated to direct bacteria killing, as the Dro gene is expressed in a variety of epithelial tissues including the trachea [38,65]. However, we do not wish to rule out a direct action of Btn on bacteria, as our in vitro conditions could have been sub-optimal for revealing an antimicrobial effect. For instance, an anionic AMP of the greater wax moth synergizes with lysozyme to kill E. coli [66], and AMPs can act synergistically in vitro through complimentary mechanisms of action [26,35,67,68]. While in vitro approaches are a powerful demonstration for AMP function, we are realizing more and more that this is not sufficient to understand peptide activity in vivo. For example, the activity of azithromycin antibiotic changes 64-fold if tested in standard in vitro conditions or with the addition of human serum [69]. In Tenebrio beetles, the AMP Tenecin-2 lacks activity against Staphylococcus aureus in vitro, but knock down via intrathoracic injection of double stranded RNA causes a significant mortality to S. aureus infection in vivo [70]. In Drosophila, Bomanin peptides do not display activity in vitro, but Bomanin-deficient haemolymph loses Candida-killing activity [21]. While AMPs were first identified for their potent microbicidal activity in vitro [4,9,71], recent studies in Drosophila have recovered striking specificity of AMPs in defence in vivo that was never predicted from in vitro analyses [6,18,19]. These results suggest both in vitro and in vivo approaches are necessary to shed light on host defence peptide activity.
It is striking that the Threonine/Alanine polymorphism in Buletin affects the fly defence against P. burhodogranariea. This polymorphism is found in wild populations of D. melanogaster, and at high frequencies in the Drosophila Genetic Reference Panel: 29% Threonine, 64% Alanine, 7% unknown at DGRP allele 2R_10633648_SNP [31,72]. A polymorphism in Diptericin A causes a profound susceptibility to defence against P. rettgeri [19], and similar polymorphisms are found in various AMP genes of flies [41,42] and other AMP genes from animals including fishes, birds, and humans [56,73,74]. We now add our study on Buletin and P. burhodogranariea to the building evidence that such polymorphisms can have major impacts on microbial control. In other species, AMP polymorphisms could have important implications on immune competence of individuals or groups. For instance: we might wonder if inbreeding in honeybees could have fixed disadvantageous AMP alleles contributing to colony collapse disorder [75]. Reduced AMP expression is also associated with conditions like psoriasis [76] or susceptibility to Pseudomonas aeruginosa infections [77,78]. A targeted screen has even suggested polymorphisms in human ß-Defensins correlate with atopic dermatitis [79]. Could polymorphisms in human AMPs help explain predisposition to similar infectious syndromes?
4. Conclusion
By uncovering a novel host defence peptide, our study contributes to a growing body of literature establishing the Drosophila systemic infection model as boasting the unique ability to reveal specific interplay of host effector-pathogen interactions. This mode of infection allows the use of the fly haemolymph as an arena to monitor pathogen growth in the presence of effectors, with fly survival as a rapid readout. While previous studies in vitro have suggested fly AMPs had generalist activities, use of specific mutations affecting individual AMP genes has now revealed specific relationships between host and pathogen. Early in vitro studies would never have predicted the highly specific requirement for only single peptides in defence against specific pathogens. Taking lessons from the fly, it should be of significant interest to characterize the differential activity of AMP polymorphisms in humans and other animals, which could reveal critical risk factors for infectious diseases.
5. Material and methods
(a) . Fly genetics
Genetic variants were isogenized into the DrosDel isogenic background over seven generations as described in [40]. The specific mutations studied here were sourced as follows: the DroSK3 mutation was generated by CRISPR-Cas9 via gRNA injection as described in [80]. The DroSK3 sequence was validated by Sanger sequencing and the nucleotide and translated sequence is shown in the electronic supplementary material, figure S3A. DroSK3 flies encode a truncated version of the Drc peptide lacking its critical Threonine needed for O-glycosylation, and we could detect variants of this truncated Drc peptide in MALDI-TOF spectra with variable degradation of the N-terminus (electronic supplementary material, figure S3A-B). The BtnThr allele used in this study was originally detected in DefSK3 flies from Parvy et al. [81] by virtue of mutation-specific MALDI-TOF proteomics while screening for possible source genes of IM7. After isogenization, iso BtnThr flies were confirmed to have a wild-type Defensin gene by polymerase chain reaction. Sequence comparisons were made using Geneious R10.
(b) . Microbe culturing conditions for infections
Bacteria were grown to mid-log phase shaking at 200 r.p.m. in their respective growth media (Luria Bertani, MRS + Mannitol) and temperature conditions, and then pelleted by centrifugation to concentrate microbes. Resulting cultures were diluted to the desired optical density (OD) at 600 nm for survival experiments, which is indicated in each figure. The following microbes were grown at 37 °C: E. coli strain 1106 (LB), P. rettgeri (LB). The following microbes were grown at 29°C: P. burhodogranariea (LB) and Acetobacter sp. ML04.1 (MRS + Mannitol).
(c) . In vitro antibacterial assays
Both the BtnThr and BtnAla versions of the 22-residue IM7 peptide were synthesized by GenicBio to a purity of greater than 95%, and silk moth Cecropin A was provided by Sigma-Aldrich at a purity of greater than or equal to 97%. Peptide preparations were verified by high performance liquid chromatography. Peptides were dissolved in water, and concentrations verified by a combination of bicinchoninic acid assay and Nanodrop A205 readings alongside a bovine serum albumin standard curve. We screened Btn for activity against both P. burhodogranariea and E. coli alone at 100 µM–1 mM, or at 100 µM in combination with serially diluted Cecropin concentrations spanning the Cecropin MIC (10 µM–0.1 µM). Microbes were allowed to grow to log-growth phase, at which point they were diluted to OD = 0.0005 in LB, and then 80 µl of this dilute culture was added to 20 µl of water or peptide mix to reach desired concentrations in a 96-well plate. Bacteria-peptide solutions were left overnight at room temperature and checked for growth the next morning, and in one experiment OD at 600 nm was recorded every 10 min using a TECAN plate reader (electronic supplementary material, figure S4).
Using these conditions, we found a minimum inhibitory concentration (MIC) for Cecropin A against E. coli 1106 of approximately 1 µM, agreeing with previous E. coli literature [82]. We found an MIC of Cecropin A against P. burhodogranariea of approximately 5 µM, though even 0.63 µM delays growth by approximately 3 h compared to no-peptide controls (electronic supplementary material, figure S4). Even at 1 mM, neither the BtnThr nor BtnAla showed any growth inhibition alone, and 100 µM peptide combinations with Cecropin A showed no reduction of MIC over Cecropin A alone. 100 µM represents the upper limit of AMP concentration in fly haemolymph after infection [83], and the concentration of Btn in vivo is probably much lower than this based on MALDI-TOF relative peak intensities [6,20,30,32]. As we tested Btn alone at 1 mM, and at 100 µM Btn + Cecropin across the Cecropin MIC range, we find that at least in our conditions using LB as diluent, Btn does not display in vitro activity.
(d) . Survival experiments
Survival experiments were performed as previously described [6], with 20 flies per vial with total replicate experiment number reported within figures (nexp). Approximately 5-day old males were used in experiments, pricked in the thorax at the pleural sulcus. Flies were flipped thrice weekly. Statistical analyses were performed using a Cox proportional hazards (CoxPH) model in R 3.6.3.
(e) . Proteomic analyses
Raw haemolymph samples were collected from immune-challenged flies for MALDI-TOF proteomic analysis as described previously [6,30]. In brief, haemolymph was collected by capillary and transferred to 0.1% trifluoroacetic acid before addition to the acetonitrile universal matrix. Representative spectra are shown. Peaks were identified via corresponding m/z values from previous studies [20,32]. Spectra were visualized using mMass, and figures were additionally prepared using Inkscape v. 0.92.
Acknowledgements
We would like to thank Adrien Schmid and Jonathan Pittet of the EPFL Proteomics Core Facility (PCF) for their technical expertise.
Contributor Information
M. A. Hanson, Email: mark.hanson@epfl.ch.
B. Lemaitre, Email: bruno.lemaitre@epfl.ch.
Data accessibility
All data are available within the manuscript and the electronic supplementary material [84].
Authors' contributions
M.A.H.: conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; S.K.: resources; B.L.: funding acquisition, investigation, project administration, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This research was supported by Sinergia grant CRSII5_186397 and Novartis Foundation 532114 awarded to B.L.
References
- 1.Hanson MA, Lemaitre B. 2020. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 62, 22-30. ( 10.1016/j.coi.2019.11.008) [DOI] [PubMed] [Google Scholar]
- 2.Lazzaro BP, Zasloff M, Rolff J. 2020. Antimicrobial peptides: application informed by evolution. Science 368, eaau5480. ( 10.1126/science.aau5480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin SJH, Cohen LB, Wasserman SA. 2020. Effector specificity and function in Drosophila innate immunity: getting AMPed and dropping Boms. PLoS Pathog. 16, e1008480. ( 10.1371/journal.ppat.1008480) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imler J-L, Bulet P. 2005. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem. Immunol. Allergy 86, 1-21. ( 10.1159/000086648) [DOI] [PubMed] [Google Scholar]
- 5.Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ. 2020. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov.19, 311-332. ( 10.1038/s41573-019-0058-8) [DOI] [PubMed] [Google Scholar]
- 6.Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B. 2019. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341. ( 10.7554/elife.44341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hultmark D, Engström A, Andersson K, Steiner H, Bennich H, Boman HG. 1983. Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. The EMBO Journal. ( 10.1002/j.1460-2075.1983.tb01465.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolff J, Schmid-Hempel P. 2016. Perspectives on the evolutionary ecology of arthropod antimicrobial peptides. Phil. Trans. R. Soc. B 371, 20150297. ( 10.1098/rstb.2015.0297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steiner H, Hultmark D, Engström AA, Bennich H, Boman HG. 1981. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292, 246-248. ( 10.1038/292246a0) [DOI] [PubMed] [Google Scholar]
- 10.Buchon N, Silverman N, Cherry S. 2014. Immunity in Drosophila melanogaster - from microbial recognition to whole-organism physiology. Nature Reviews Immunol. 14, 796-810. ( 10.1038/nri3763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemaitre B, Hoffmann J. 2007. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697-743. ( 10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- 12.Kaneko T et al. 2004. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637-649. ( 10.1016/s1074-7613(04)00104-9) [DOI] [PubMed] [Google Scholar]
- 13.Leulier F et al. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 4, 478-484. ( 10.1038/ni922) [DOI] [PubMed] [Google Scholar]
- 14.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21, 2568-2579. ( 10.1093/emboj/21.11.2568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemaitre B, Reichhart JM, Hoffmann JA. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl Acad. Sci. USA 94, 14 614-14 619. ( 10.1073/pnas.94.26.14614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanji T, Hu X, Weber ANR, Ip YT. 2007. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila melanogaster. Molecular and Cellular Biology 27, 4578-4588. ( 10.1128/MCB.01814-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemmons AW, Lindsay SA, Wasserman SA. 2015. An effector peptide family required for Drosophila toll-mediated immunity. PLoS Pathog. 11, e1004876. ( 10.1371/journal.ppat.1004876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen LB, Lindsay SA, Xu Y, Lin SJH, Wasserman SA. 2020. The Daisho peptides mediate Drosophila defense against a subset of filamentous fungi. Front Immunol. 11, 9. ( 10.3389/fimmu.2020.00009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unckless RL, Howick VM, Lazzaro BP. 2016. Convergent balancing selection on an antimicrobial peptide in Drosophila. Curr. Biol. 26, 257-262. ( 10.1016/j.cub.2015.11.063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanson MA, Cohen LB, Marra A, Iatsenko I, Wasserman SA, Lemaitre B. 2021. The Drosophila Baramicin polypeptide gene protects against fungal infection. PLoS Pathog. 17, e1009846. ( 10.1371/journal.ppat.1009846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindsay SA, Lin SJH, Wasserman SA. 2018. Short-form bomanins mediate humoral immunity in Drosophila. J. Innate Immun. 10, 306-314. ( 10.1159/000489831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulet P, Urge L, Ohresser S, Hetru C, Otvos L. 1996. Enlarged scale chemical synthesis and range of activity of drosocin, an O-glycosylated antibacterial peptide of Drosophila. Eur. J. Biochem. 238, 64-69. ( 10.1111/j.1432-1033.1996.0064q.x) [DOI] [PubMed] [Google Scholar]
- 23.Carboni A, Hanson MA, Lindsay SA, Wasserman SA, Lemaitre B. 2021. Cecropins contribute to Drosophila host defence against fungal and Gram-negative bacterial infection. Genetics. iyab188. ( 10.1093/genetics/iyab188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casteels-Josson K, Capaci T, Casteels P, Tempst P. 1993. Apidaecin multipeptide precursor structure: a putative mechanism for amplification of the insect antibacterial response. The EMBO J. 12, 1569-78. ( 10.1002/j.1460-2075.1993.tb05801.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedengren M, Borge K, Hultmark D. 2000. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem. Biophys. Res. 279, 574-581. ( 10.1006/bbrc.2000.3988) [DOI] [PubMed] [Google Scholar]
- 26.Rabel D, Charlet M, Ehret-Sabatier L, Cavicchioli L, Cudic M, Otvos L, Bulet P. 2004. Primary structure and in vitro antibacterial properties of the Drosophila melanogaster attacin C pro-domain. J. Biol. Chem. 279, 14 853-14 859. ( 10.1074/jbc.M313608200) [DOI] [PubMed] [Google Scholar]
- 27.Huang J et al. 2020. The BaramicinA gene is required at several steps of the host defense against Enterococcus faecalis and Metarhizium robertsii in a septic wound infection model in Drosophila melanogaster. bioRxiv; 11. ( 10.1101/2020.11.23.394809) [DOI] [Google Scholar]
- 28.Lezi E et al. 2018. An antimicrobial peptide and its neuronal receptor regulate dendrite degeneration in aging and infection. Neuron 97, 125-138.e5. ( 10.1016/j.neuron.2017.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.UniProt Consortium. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480-D489. ( 10.1093/nar/gkaa1100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. 1998. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc. Natl Acad. Sci. USA 95, 11 342-11 347. ( 10.1073/pnas.95.19.11342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazzaro BP, Clark AG. 2003. Molecular population genetics of inducible antibacterial peptide genes in Drosophila melanogaster. Mol. Biol. Evol. 20, 914-923. ( 10.1093/molbev/msg109) [DOI] [PubMed] [Google Scholar]
- 32.Levy F, Rabel D, Charlet M, Bulet P, Hoffmann JA, Ehret-Sabatier L. 2004. Peptidomic and proteomic analyses of the systemic immune response of Drosophila. Biochimie. 86, 607-616. ( 10.1016/j.biochi.2004.07.007) [DOI] [PubMed] [Google Scholar]
- 33.Kragol G, Lovas S, Varadi G, Condie BA, Hoffmann R, Otvos L. 2001. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 40, 3016-3026. ( 10.1021/bi002656a) [DOI] [PubMed] [Google Scholar]
- 34.Otvos LOI et al. 2000. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 39, 14 150-14 159. ( 10.1021/bi0012843) [DOI] [PubMed] [Google Scholar]
- 35.Rahnamaeian M, Cytryńska M, Zdybicka-Barabas A, Vilcinskas A. 2016. The functional interaction between abaecin and pore-forming peptides indicates a general mechanism of antibacterial potentiation. Peptides 78, 17-23. ( 10.1016/j.peptides.2016.01.016) [DOI] [PubMed] [Google Scholar]
- 36.Bikker FJ, Kaman-van Zanten WE, de Ruit A-MBC dV-v, Voskamp-Visser I, van Hooft PAV, Mars- Groenendijk RH, de Visser PC, Noort D. 2006. Evaluation of the antibacterial spectrum of drosocin analogues. Chem. Biol. Drug Des. 68, 148-153. ( 10.1111/j.1747-0285.2006.00424.x) [DOI] [PubMed] [Google Scholar]
- 37.Bulet P et al. 1993. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J. Biol. Chem. 268, 14 893-14 897. [PubMed] [Google Scholar]
- 38.Sanchez Bosch P et al. 2019. Adult Drosophila lack hematopoiesis but rely on a blood cell reservoir at the respiratory epithelia to relay infection signals to surrounding tissues. Dev. Cell 51, 1-17. ( 10.1016/j.devcel.2019.10.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marra A, Hanson MA, Kondo S, Erkosar B, Lemaitre B. In press. Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. Microbiology ( 10.1101/2021.03.19.436153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira ÁG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L. 2014. The Toll-Dorsal pathway is required for resistance to viral oral infection in Drosophila. PLoS Pathog. 10, e1004507. ( 10.1371/journal.ppat.1004507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chapman JR, Hill T, Unckless RL. 2019. Balancing selection drives maintenance of genetic variation in Drosophila antimicrobial peptides. Genome Biol. Evol. 11, 2691-2701. ( 10.1093/gbe/evz191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unckless RL, Lazzaro BP. 2016. The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Phil. Trans. R. Soc. B. 371, 20150291. ( 10.1098/rstb.2015.0291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juneja P, Lazzaro BP. 2009. Providencia sneebia sp. nov. and Providencia burhodogranariea sp. nov., isolated from wild Drosophila melanogaster. Int. J. Syst. Evol. Microbiol. 59, 1108-1111. ( 10.1099/ijs.0.000117-0) [DOI] [PubMed] [Google Scholar]
- 44.Karlsson C, Korayem AM, Scherfer C, Loseva O, Dushay MS, Theopold U. 2004. Proteomic analysis of the Drosophila larval hemolymph clot. J. Biol. Chem. 279, 52 033-52 041. ( 10.1074/jbc.M408220200) [DOI] [PubMed] [Google Scholar]
- 45.Waumans Y, Baerts L, Kehoe K, Lambeir A-M, De Meester I. 2015. The dipeptidyl peptidase family, prolyl oligopeptidase, and prolyl carboxypeptidase in the immune system and inflammatory disease, including atherosclerosis. Front. Immunol. 6 ( 10.3389/fimmu.2015.00387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson MA, Hamilton PT, Perlman SJ. 2016. Immune genes and divergent antimicrobial peptides in flies of the subgenus Drosophila. BMC Evol. Biol. 16, 228. ( 10.1186/s12862-016-0805-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanson MA, Lemaitre B, Unckless RL. 2019. Dynamic evolution of antimicrobial peptides underscores trade-offs between immunity and ecological fitness. Front. Immunol. 10, 2620. ( 10.3389/fimmu.2019.02620) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W et al. 2014. Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. ( 10.1007/s00726-014-1820-1) [DOI] [PubMed] [Google Scholar]
- 49.Bulmer MS, Crozier RH. 2004. Duplication and diversifying selection among termite antifungal peptides. Mol. Biol. Evol. 21, 2256-2264. ( 10.1093/molbev/msh236) [DOI] [PubMed] [Google Scholar]
- 50.Kraaijeveld AR, Van Alphen JJ, Godfray HC. 1998. The coevolution of host resistance and parasitoid virulence. Parasitology 116, S29-S45. ( 10.1017/s0031182000084924) [DOI] [PubMed] [Google Scholar]
- 51.Leitão AB et al. 2020. Constitutive activation of cellular immunity underlies the evolution of resistance to infection in Drosophila. eLife 9, e59095. ( 10.7554/eLife.59095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramond E et al. 2020. The adipokine NimrodB5 regulates peripheral hematopoiesis in Drosophila. FEBS J. 287, 3399-3426. ( 10.1111/febs.15237) [DOI] [PubMed] [Google Scholar]
- 53.Belmonte RL, Corbally M-K, Duneau DF, Regan JC. 2019. Sexual dimorphisms in innate immunity and responses to infection in Drosophila melanogaster. Front. Immunol. 10, 3075. ( 10.3389/fimmu.2019.03075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta V, Frank AM, Matolka N, Lazzaro BP. 2022. Inherent constraints on a polyfunctional tissue lead to a reproduction-immunity tradeoff. BMC Biol. 20, 127. ( 10.1186/s12915-022-01328-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwenke RA, Lazzaro BP, Wolfner MF. 2015. Reproduction–Immunity Trade-Offs in Insects. Annu. Rev. Entomol. 61, 239-256. ( 10.1146/annurev-ento-010715-023924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hollox EJ, Armour JAL. 2008. Directional and balancing selection in human beta-defensins. BMC Evol. Biol. 8. ( 10.1186/1471-2148-8-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tennessen JA, Blouin MS. 2008. Balancing selection at a frog antimicrobial peptide locus: fluctuating immune effector alleles? Mol. Biol. Evol. 25, 2669-2680. ( 10.1093/molbev/msn208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox CR, Gilmore MS. 2007. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infection and Immunity 75, 1565-1576. ( 10.1128/IAI.01496-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barajas-azpeleta R, Wu J, Gill J, Welte R. 2018. Antimicrobial peptides modulate long-term memory. PLoS Genetics 1-26. ( 10.1371/journal.pgen.1007440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ebrahim SAM, Talross GJS, Carlson JR. 2021. Sight of parasitoid wasps accelerates sexual behavior and upregulates a micropeptide gene in Drosophila. Nat Commun. 12, 2453. ( 10.1038/s41467-021-22712-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobler JM, Rodriguez Jimenez FJ, Petcu I, Grunwald Kadow IC. 2020. Immune receptor signaling and the mushroom body mediate post-ingestion pathogen avoidance. Curr. Biol. 30 4693-4709. ( 10.1016/j.cub.2020.09.022) [DOI] [PubMed] [Google Scholar]
- 62.Sinner MP, Masurat F, Ewbank JJ, Pujol N, Bringmann H. 2021. Innate immunity promotes sleep through epidermal antimicrobial peptides. Curr. Biol. 31, 564-577. ( 10.1016/j.cub.2020.10.076) [DOI] [PubMed] [Google Scholar]
- 63.Jumper J et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583-589. ( 10.1038/s41586-021-03819-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols 3, 163-175. ( 10.1038/nprot.2007.521) [DOI] [PubMed] [Google Scholar]
- 65.Tzou P et al. 2000. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 13, 737-748. ( 10.1016/S1074-7613(00)00072-8) [DOI] [PubMed] [Google Scholar]
- 66.Zdybicka-Barabas A et al. 2012. Synergistic action of Galleria mellonella anionic peptide 2 and lysozyme against Gram-negative bacteria. Biochim Biophys Acta. 1818, 2623-2635. ( 10.1016/j.bbamem.2012.06.008) [DOI] [PubMed] [Google Scholar]
- 67.Peng S et al. 2018. Mechanism of actions of Oncocin, a proline-rich antimicrobial peptide, in early elongation revealed by single-molecule FRET. Protein Cell 9, 890-895. ( 10.1007/s13238-017-0495-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu G, Baeder DY, Regoes RR, Rolff J. 2016. The more the better? Combination effects of antimicrobial peptides. Antimicrobial Agents and Chemotherapy. AAC.02434-15. ( 10.1128/AAC.02434-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Belanger CR, Lee AH-Y, Pletzer D, Dhillon BK, Falsafi R, Hancock REW. 2020. Identification of novel targets of azithromycin activity against Pseudomonas aeruginosa grown in physiologically relevant media. Proc. Natl Acad. Sci. USA 117, 33 519-33 529. ( 10.1073/pnas.2007626117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanchi C, Johnston PR, Rolff J. 2017. Evolution of defence cocktails: Antimicrobial peptide combinations reduce mortality and persistent infection. Mol. Ecol. 26, 5334-5343. ( 10.1111/mec.14267) [DOI] [PubMed] [Google Scholar]
- 71.Zasloff M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl Acad. Sci. USA 84, 5449-5453. ( 10.1073/pnas.84.15.5449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mackay TFC et al. 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482, 173-178. ( 10.1038/nature10811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hellgren O, Sheldon BC, Buckling A. 2010. In vitro tests of natural allelic variation of innate immune genes (avian β-defensins) reveal functional differences in microbial inhibition. J. Evol. Biol. 23, 2726-2730. ( 10.1111/j.1420-9101.2010.02115.x) [DOI] [PubMed] [Google Scholar]
- 74.Hellgren O, Sheldon BC. 2011. Locus-specific protocol for nine different innate immune genes (antimicrobial peptides: β-defensins) across passerine bird species reveals within-species coding variation and a case of trans-species polymorphisms. Mol. Ecol. Resour. 11, 686-692. ( 10.1111/j.1755-0998.2011.02995.x) [DOI] [PubMed] [Google Scholar]
- 75.Fürst MA, McMahon DP, Osborne JL, Paxton RJ, Brown MJF. 2014. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506, 364-366. ( 10.1038/nature12977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marcinkiewicz M, Majewski S. 2016. The role of antimicrobial peptides in chronic inflammatory skin diseases. Adv. Dermatol. Allergol./Postępy Dermatol. Alergol. 33, 6-12. ( 10.5114/pdia.2015.48066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bergsson G et al. 2009. LL-37 Complexation with Glycosaminoglycans in Cystic Fibrosis Lungs Inhibits Antimicrobial Activity, Which Can Be Restored by Hypertonic Saline. J. Immunol. 183, 543-551. ( 10.4049/jimmunol.0803959) [DOI] [PubMed] [Google Scholar]
- 78.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. 1997. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 88, 553-560. ( 10.1016/S0092-8674(00)81895-4) [DOI] [PubMed] [Google Scholar]
- 79.Prado-Montes de Oca E, García-Vargas A, Lozano-Inocencio R, Gallegos-Arreola MP, Sandoval-Ramírez L, Dávalos-Rodríguez NO, Figuera LE. 2007. Association of β-Defensin 1 single nucleotide polymorphisms with atopic dermatitis. Int. Arch. Allergy Immunol. 142, 211-218. ( 10.1159/000097023) [DOI] [PubMed] [Google Scholar]
- 80.Kondo S, Ueda R. 2013. Highly Improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 195, 715-721. ( 10.1534/genetics.113.156737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parvy J-P et al. 2019. The antimicrobial peptide defensin cooperates with tumour necrosis factor to drive tumour cell death in Drosophila. eLife 8, e45061. ( 10.7554/eLife.45061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silvestro L, Weiser JN, Axelsen PH. 2000. Antibacterial and antimembrane activities of cecropin A in Escherichia coli. Antimicrob Agents Chemother. 44, 602-607. ( 10.1128/AAC.44.3.602-607.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fehlbaum P et al. 1994. Insect immunity: Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 269, 33 159-33 163. [PubMed] [Google Scholar]
- 84.Hanson MA, Kondo S, Lemaitre B. 2022. Data from: Drosophila immunity: the Drosocin gene encodes two host defence peptides with pathogen-specific roles. Figshare. ( 10.6084/m9.figshare.c.6035718) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available within the manuscript and the electronic supplementary material [84].