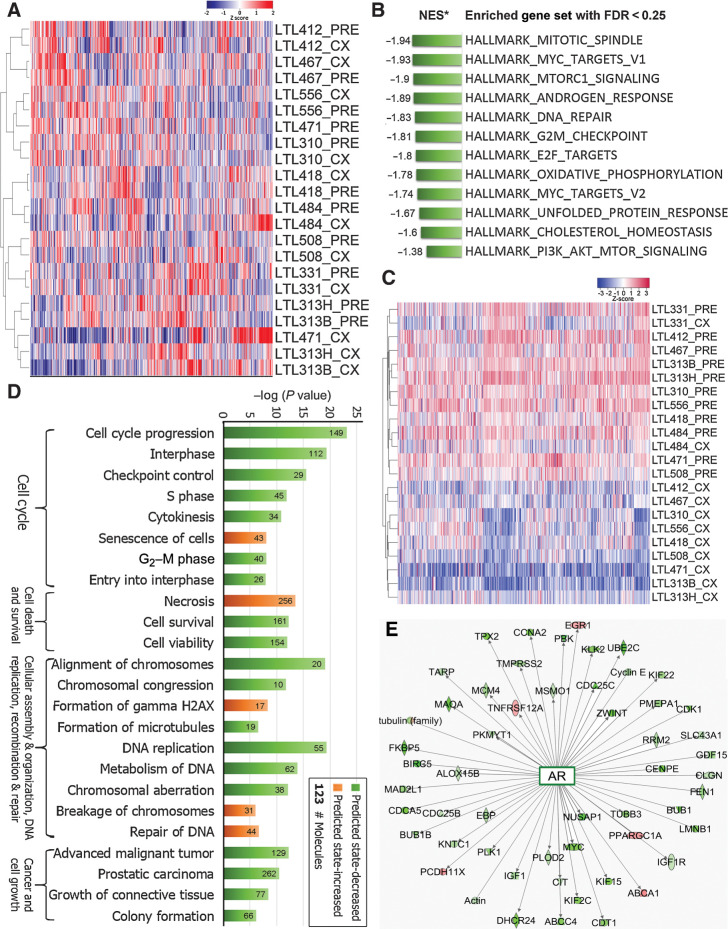
Figure 2.
Compared with the precastration PDXs, dormant PDXs showed specific changes in gene expressions. A, genome-wide unsupervised hierarchical cluster showed most dormant (CX) PDXs clustered with their parental precastration (PRE) PDXs. B, GSEA based on whole transcripts without preranking identified 12 significantly enriched gene sets in PRE PDXs (FDR < 0.25). C, Unsupervised hierarchical clustering based on the Leading Edge genes generated by GSEA showed a distinct separation between PRE and most CX PDXs. D, IPA revealed that deregulated pathways were mainly associated with cell cycle, cell death and survival, cellular assembly, DNA repair, and cell growth. E, Fifty-three of 58 differentially-expressed genes that were indicated by IPA to have a direct interaction with AR were downregulated in PDXs of dormant prostate cancer.