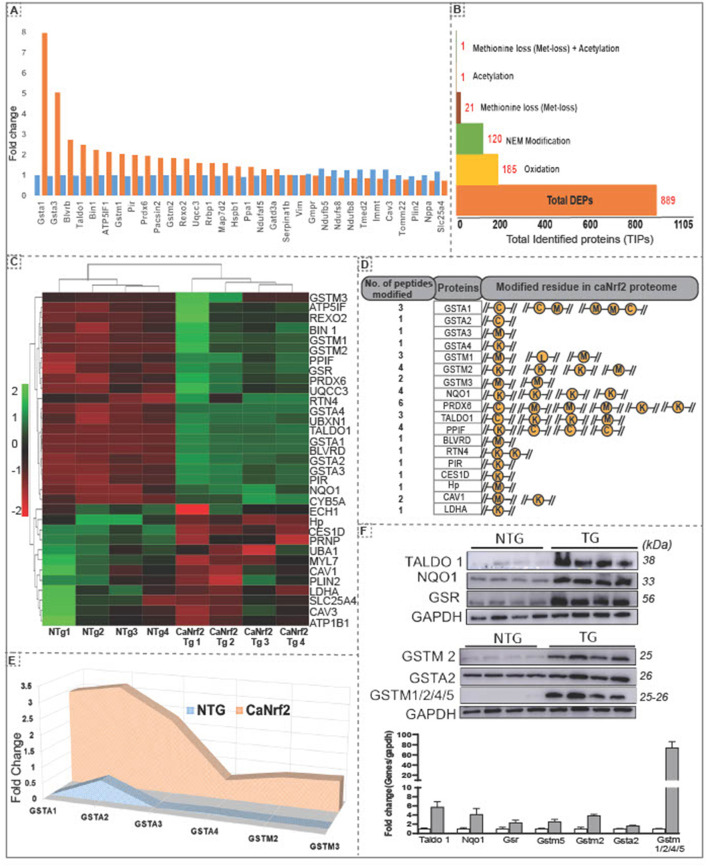
Figure 2.
Hierarchical clustering of differentially expressed proteins and amino acid modifications in proteins in the RS hearts. (A) Unsupervised clustering of total identified proteins showed 32 DEPs at 2 fold change with Gsta1, Gsta2, Blvrd, Taldo1, Bin1, Atp5lf1, gstm1,Pir and Prdx6 as the top upregulated proteins in CaNrf2 hearts. (B) Spectrum view by proteome scaffold software identified different types of modification in differentially expressed proteins (889) with 185 proteins having oxidation,120 with NEM modification, 21 with loss of methionine peptide, 1 with acetylation, 1 with acetylation and 1 peptide with both loss of methylation and acetylation. (C) CaNrf2 proteome heat map using R studio for highly upregulated proteins (based on log2 FC) identified in scaffold software. (D) Highly upregulated proteins are associated with amino acid modifications in peptide(s). Data showing the number of peptides modified in each protein and modified residue in each peptide. A representative peptide is shown in yellow color. (E) TMT proteome identified GSR family of proteins as top enriched in CaNrf2 hearts in comparison with NTg. Significantly changed proteins are Gsta2, Gsta3, Gsta4, Gstm2, Gstm3 and Gsta1 (based on log2 fold change ratio). (F) Immunoblot validation for the selected proteins in TG vs. NTg hearts. Corresponding densitometry analysis using Image-J is shown.