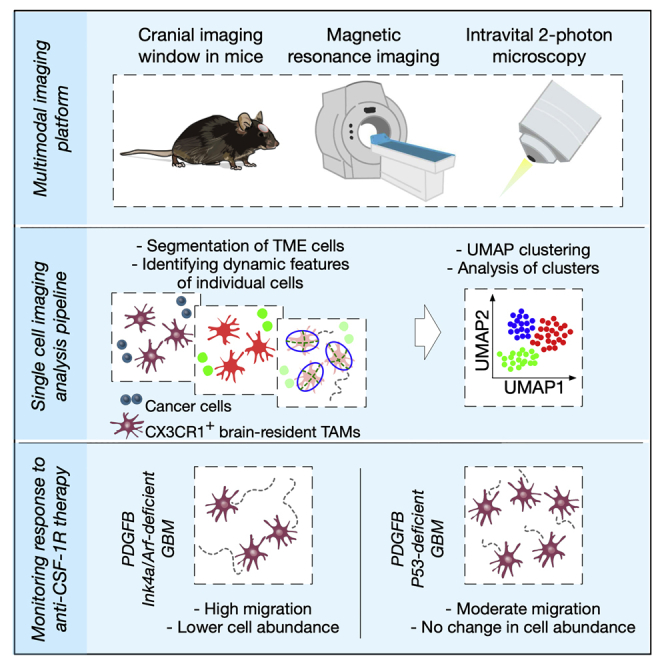
Summary
Tumors evolve in a dynamic communication with their native tissue environment and recruited immune cells. The diverse components of the tumor microenvironment (TME) can critically regulate tumor progression and therapeutic response. In turn, anticancer treatments may alter the composition and functions of the TME. To investigate this continuous dialog in the context of primary brain cancers, we developed a multimodal longitudinal imaging strategy. We combined macroscopical magnetic resonance imaging with subcellular resolution two-photon intravital microscopy, and leveraged the power of single-cell analysis tools to gain insights into the ongoing interactions between different components of the TME and cancer cells. Our experiments revealed that the migratory behavior of tumor-associated macrophages is different in genetically distinct glioblastomas, and in response to macrophage-targeted therapy. These results underscore the importance of studying cancer longitudinally in an in vivo setting, to reveal complex and dynamic alterations in the TME during disease progression and therapeutic intervention.
Subject areas: Microenvironment, Medical imaging, Physics magnetic resonance imaging, Cancer
Graphical abstract
Highlights
-
•
Multimodal longitudinal imaging of glioblastomas by MRI and two-photon microscopy
-
•
Single-cell imaging analysis to investigate dynamics of the tumor microenvironment
-
•
Diverse abundance and behavior of macrophages in genetically distinct glioblastomas
Microenvironment; Medical imaging; Physics magnetic resonance imaging; Cancer
Introduction
Cancers develop and progress within complex environments, involving multiple interactions with the surrounding healthy tissue along with newly infiltrating immune cells, which collectively contribute to the regulation of tumor progression and metastasis (Quail and Joyce, 2013). The interaction between malignant and non-malignant cells is constantly evolving, impacting disease progression and response to treatment (Klemm and Joyce, 2015). Given the increasing application of immunomodulatory drugs for the treatment of patients with cancer, it is crucial to gain a deeper understanding into dynamically evolving processes within the tumor microenvironment (TME), for example, to determine the optimal therapeutic window for specific anticancer treatments, to understand how different genetic mutations impact tumor progression, and to reveal mechanisms of acquired or intrinsic resistance (Bejarano et al., 2021; Ott et al., 2021).
In glioblastoma, the most malignant form of primary brain tumor in adults (Stupp et al., 2005), the TME is substantially different compared to the TME of other cancer types. There are several unique properties of the normal brain including the presence of brain-resident cell types such as microglia, astrocytes and neurons, a distinctive extracellular matrix (ECM) composition, and the presence of the blood–brain barrier (BBB), protecting the brain from systemic inflammation (Quail and Joyce, 2017). However, in advanced brain malignancies, the BBB is often compromised, enabling infiltration of immune cells from the circulation (Arvanitis et al., 2020). Indeed, different types of peripheral immune cells have been found in human gliomas, and interestingly, the relative abundance of tissue-resident myeloid cells, recruited myeloid cells, and lymphoid cells was shown to be associated with the mutational status of the metabolic enzyme isocitrate dehydrogenase 1 (IDH1) (Friebel et al., 2020; Klemm et al., 2020), a mutation that generally discriminates low-grade (IDH1 mutant) and high-grade (IDH1 wild-type) gliomas (glioblastoma; GBM). In addition to the mutational status of gliomas, the TME has also been shown to be altered as a consequence of standard-of-care radiotherapy (Akkari et al., 2020; Magod et al., 2021). However, it is currently unknown whether the TME of GBM evolves dynamically over the course of disease progression, and in response to treatment, both in terms of abundance and behavior of specific TME components.
To capture microenvironmental changes that occur in a gradual and dynamic manner, we developed a multimodal longitudinal imaging approach for GBM that combines a translationally relevant diagnostic technique, magnetic resonance imaging (MRI) (Smits, 2021), with the power of high-resolution two-photon intravital microscopy (IVM) (Suijkerbuijk and van Rheenen, 2017). Longitudinal in vivo imaging using two-photon IVM has been used to study cellular dynamics in brain tumors, including cancer stem cell-driven GBM growth (Lathia et al., 2011) and the development of functional multicellular network structures between individual astrocytoma cells (Osswald et al., 2015). Here, we sought to specifically analyze the ongoing interactions between different components of the TME and cancer cells in genetically distinct GBMs. An increasing body of evidence shows that the glioma TME can fundamentally change during disease progression and in response to treatment (Akkari et al., 2020; Magod et al., 2021). As such, the combination of MRI and two-photon IVM that we use herein enables one to integrate measurements of the overall tumor growth and/or regression with the composition and behavior of the TME at a single-cell level. This powerful multimodal strategy allowed us to study GBM at both a macroscopical and microscopical level, respectively. Moreover, we implemented existing data science tools to perform single-cell imaging analysis of complex and large IVM datasets, containing information from thousands of individual cells that were imaged in the same individuals over many weeks, thereby enabling an unbiased interrogation of the changing TME.
Results
Multimodal imaging pipeline enables the long-term longitudinal imaging of glioblastomas
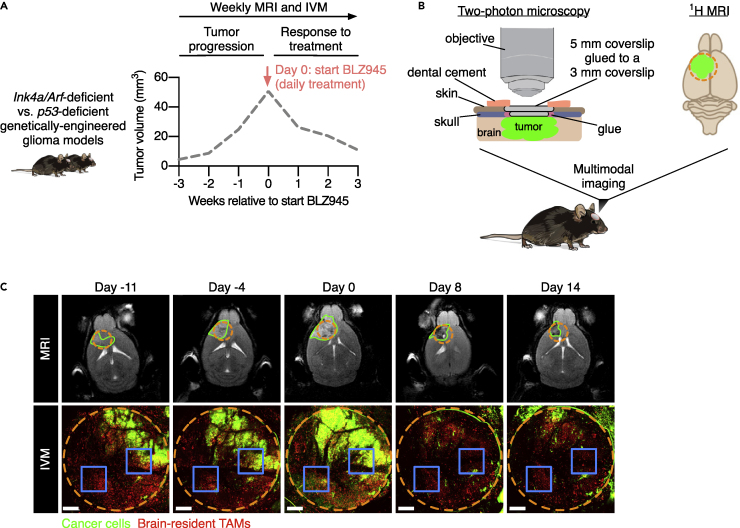
To image brain tumors during disease progression and in response to treatment in long-term multimodal MRI-IVM experiments (Figure 1A), we developed a metal-free cranial imaging window (CIW) setup that is compatible with MRI (Figures 1B and S1). The CIW was surgically implanted in 4- to 7-week-old mice (Figure S2A), and platelet-derived growth factor-β (PDGF-β)-driven GBMs were simultaneously induced (Hambardzumyan et al., 2009). First, a 3-mm diameter craniotomy was made using a biopsy punch and the bone flap was evaluated for the presence of dura mater residuals to confirm that the dura mater and the native environment of the brain remained intact. Subsequently, the site of the craniotomy was sealed with two coverslips that are glued together: one of 3-mm diameter that completely covers the craniotomy and prevents skull regrowth, and one of 5-mm diameter that was used to fix the CIW to the skull. Lastly, a metal-free head bar was placed for the fixation of the mouse under the two-photon microscope and to position the CIW perpendicular to the objective, and the exposed skull was sealed using dental cement. Importantly, the surgical procedure minimally impacted animal welfare, and all the mice had completely recovered before starting to develop tumors 3.5 weeks after induction (Figures S2B and S2C). Together, our multimodal MRI-IVM pipeline allows the imaging of mice over several weeks, and shows the potential to interrogate cells of interest in their natural tissue environment.
Figure 1.
Multimodal IVM-MRI imaging to visualize dynamics of the brain TME over multiple weeks
(A) Schematic showing the experimental design used to investigate TME dynamics during tumor progression and in response to BLZ945 treatment in Ink4a/Arf-deficient and p53-deficient GBMs. Mice were imaged weekly to assess tumor growth by MRI and to monitor TME dynamics by IVM. The dashed gray line indicates a representative tumor growth curve of a GBM-bearing mouse.
(B) In order to obtain visual access to the brain tumor and perform multimodal IVM-MRI, a CIW was surgically implanted in the mouse skull at the site of tumor induction (left). The incorporation of metal-free components enabled the concomitant use of MRI (right). The dashed orange circle indicates the position of the CIW on the top of the tumor (green).
(C) Representative example of longitudinal multimodal imaging of the same mouse by MRI and IVM before and after treatment with the CSF-1R inhibitor BLZ945 (treatment initiated at day 0). The upper MRI images show the anatomical location of the tumor (green line) and the projection of the CIW position (dashed orange circle). The lower panels show matching two-photon microscopy tile scans visualizing the dynamics of GFP-positive cancer cells (green) and brain-resident TAMs (red) through the CIW (dashed orange circle). The blue squares indicate an example of the location of two FOVs that were used to record time-lapses throughout the duration of the longitudinal IVM-MRI imaging experiment. Scale bars: 400 μm. Brain silhouette in (B) was sourced from https://scidraw.io/. See also Figures S1 and S2.
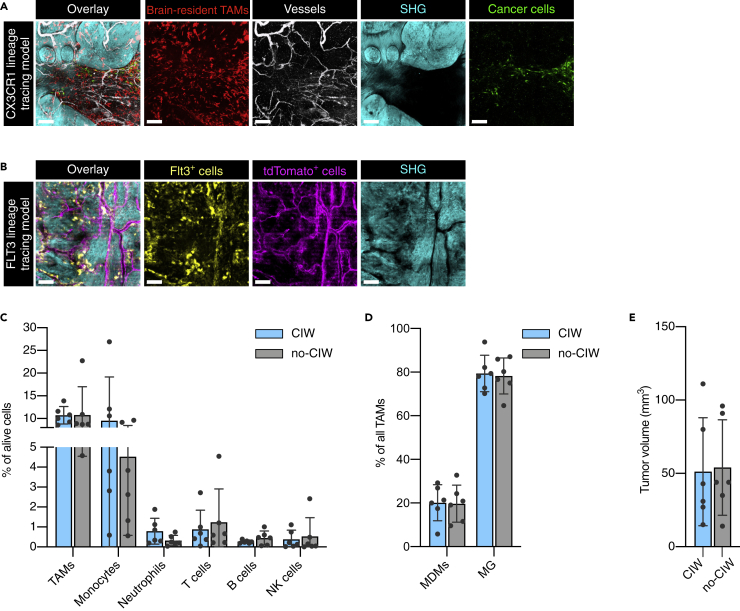
Lineage-tracing models to study the in vivo behavior of different TME components
Analysis of total immune cell infiltrates in patients with glioblastoma revealed that macrophages represent the major immune cell constituent (Friebel et al., 2020; Klemm et al., 2020), and their accumulation correlates with higher tumor grade (Komohara et al., 2008). These tumor-associated macrophages (TAMs) consist of brain-resident cells, including parenchymal microglia (MG) and non-parenchymal border-associated macrophages (BAMs), as well as peripherally recruited monocyte-derived macrophages (MDMs) (Ochocka et al., 2021). Brain-resident and recruited TAMs are morphologically distinct and display differential migratory behavior in vivo (Chen et al., 2019). In order to study the longitudinal dynamics of brain-resident TAMs and peripherally recruited immune cells in detail, we employed two different cell lineage-tracing strategies (Figures 2A and 2B) (Bowman et al., 2016). To specifically label brain-resident TAMs, we used Cx3cr1:CreERT2-IRES-YFP;Rosa26:lslTdTomato mice that express TdTomato in Cx3cr1-expressing cells (tissue-resident macrophages and circulating monocytes) upon tamoxifen injection (Parkhurst et al., 2013) (termed CX3CR1 lineage-tracing model). Three weeks after tamoxifen-induced labeling, when circulating myeloid cells have been replenished by tdTomato− cells and brain-resident macrophages remain tdTomato+ (Bowman et al., 2016; Jordao et al., 2019), we induced green fluorescent protein (GFP)+ PDGF-β-driven GBMs in order to detect cancer cells in the same model (Figure 2A). Additionally, blood vessels were imaged using fluorescently labeled dextran, and the extracellular matrix (ECM) was visualized by two-photon second harmonic generation (SHG) imaging (Figure 2A), providing complementary insights into other cellular and non-cellular components of the TME.
Figure 2.
TME characterization of lineage-tracing mouse models
(A) Representative two-photon microscopy images of the CX3CR1 lineage-tracing model showing brain-resident TAMs (red), dextran-Pacific Blue-labeled vessels (white), SHG imaging signal (cyan), and GFP-expressing cancer cells (green). Scale bars: 150 μm.
(B) The FLT3 lineage-tracing model allowed the detection of Flt3+ GFP-expressing immune cells (yellow), ectopic tdTomato mainly expressed in vessel-like structures (violet), and SHG imaging signal (cyan). Scale bars: 150 μm.
(C and D) Flow cytometry analysis showed an equivalent abundance of the major immune cell types in GBM-bearing mice with and without a CIW (paired Wilcoxon test, no statistically significant differences were observed). Each dot represents one tumor.
(E) Endpoint volume of the tumors (measured by MRI) that were characterized by flow cytometry in (C) and (D) (Mann-Whitney test, no statistically significant difference was observed). Each dot represents one tumor. Data in (C), (D), and (E) are represented as mean ± SD. See also Table S1.
By incorporating another lineage-tracing model, we also sought to investigate the contribution of recruited peripheral immune cells. We used Flt3:Cre; Rosa26:mTmG mice in which all cells of the hematopoietic lineage are labeled with membrane-localized GFP, whereas all other cells express cell membrane-localized tdTomato (termed FLT3 lineage-tracing model) (Figure 2B and Video S1) (Bowman et al., 2016; Boyer et al., 2011). We first determined whether the implantation of the CIW impacts the composition of the GBM TME or potentially induces an inflammatory response. Flow cytometry analysis of GBMs that developed in the presence or absence of a CIW showed an equivalent abundance of the major immune cell types, indicating that the CIW surgery does not evidently affect the TME composition (Figures 2C, 2D, and 2E).
Peripheral FLT3+ cells are shown in yellow and tdTomatoHIGH cells in violet. The total duration of the time-lapse is approximately 30 min with an interval of 1 min between each frame.
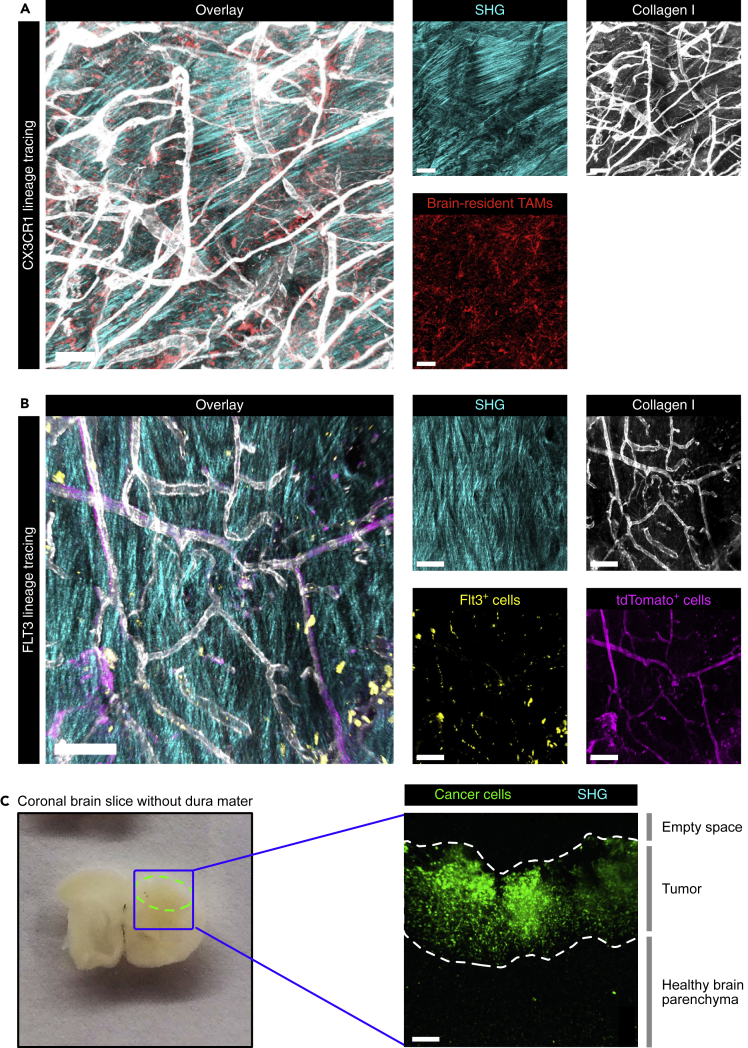
Next, we assessed whether our imaging strategy allows the detection of the meningeal membranes which are composed of fibrous tissue including collagenic and elastic fibers (Protasoni et al., 2011). The meningeal membranes lie immediately beneath the skull and include the dura, pia, and arachnoid mater. To determine whether these meningeal membranes show SHG signal, we performed ex vivo 2-photon imaging of the dura mater and of brain slices (where the pia and arachnoid mater remain attached after tissue harvesting) (Figures 3A, 3B and 3C). Interestingly, we found that the dura mater was the main source of the SHG signal (Figures 3A and 3B), while no SHG signal was observed in the brain slices that contain the other meningeal membranes (Figure 3C). Together, these results show that the MRI-IVM pipeline, in combination with cell lineage-tracing models, is a robust and powerful strategy to investigate the fate of the cells of interest in vivo. Moreover, the SHG signal can be used as a key anatomical reference to discriminate the dura mater from the other meningeal membranes and the brain parenchyma.
Figure 3.
The dura mater, but not the brain parenchyma, shows SHG imaging signal
(A and B) Representative two-photon microscopy images of the dura mater dissected from GBM-bearing (A) CX3CR1 lineage-tracing mice and (B) FLT3 lineage-tracing mice showing the second harmonic generation (SHG) signal. Scale bars: 100 μm.
(C) 425-μm thick tumor-bearing brain slices without dura mater (left, tumor area outlined by the dashed green line) imaged by two-photon microscopy (right) showed GFP-positive cancer cells and the adjacent healthy brain parenchyma, but no SHG signal. Scale bar: 100 μm. See also Table S1.
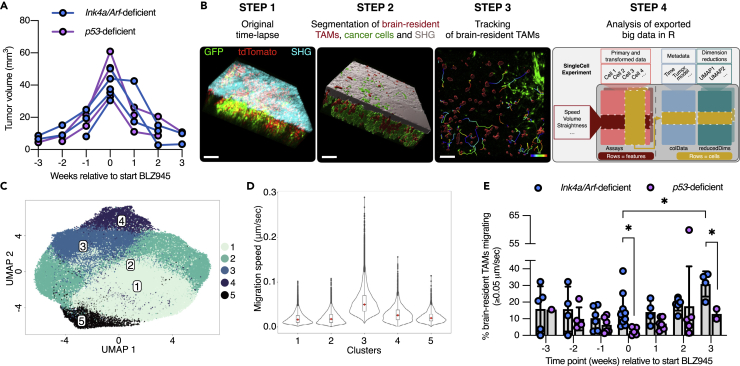
Single-cell imaging analysis of IVM data reveals different macrophage subsets during tumor progression and in response to treatment
An increasing body of evidence indicates that the TME can be differentially educated depending on specific genetic alterations in the cancer cells (Wellenstein and de Visser, 2018). However, precisely how the complex TME changes dynamically over the course of tumor progression, and in response to therapy, in genetically distinct cancers remains to be fully explored. We therefore applied our multimodal imaging platform to first investigate the dynamics of brain-resident TAMs in PDGF-β-driven GBMs that are deficient for either of the tumor suppressor genes Ink4a/Arf or p53 (Figure 1A). These tumors closely mimic the genetic profile of patient glioblastomas, in which deletion of CDKN2A (encoding for both p16INK4A and p14ARF) is observed in 57.8% of patients, and p53 mutation/deletion occurs in 27.9% of patients (Brennan et al., 2013; Hambardzumyan et al., 2009; Ozawa et al., 2014).
At 3.5 weeks after tumor initiation and CIW implantation in the CX3CR1 lineage-tracing model, we began MRI scans on a weekly basis to screen for the presence of tumors. As soon as small tumors were detected (<10 mm3), mice were subjected to weekly MRI and IVM to monitor cellular dynamics during disease progression (Figures 1A, 1C, and 4A). Subsequently, advanced GBMs (>30 mm3) were treated with BLZ945, a potent and highly selective small-molecule inhibitor of the colony-stimulating factor-1 receptor (CSF-1R), that targets macrophages specifically and depletes MG in healthy brain tissue (Pyonteck et al., 2013). We have previously shown that secreted factors in the GBM TME protect TAMs from BLZ945-mediated depletion in the PDGF-β; Ink4a/Arf-deficient model, and that these surviving TAMs then become antitumorigenic (Pyonteck et al., 2013). Consequently, we found that BLZ945 treatment regresses GBM (Figure 4A) and significantly prolongs survival in multiple murine glioma models (Akkari et al., 2020; Pyonteck et al., 2013; Quail et al., 2016).
Figure 4.
Analysis of a large IVM dataset reveals temporal dynamics of brain-resident TAMs during the treatment of GBM
(A) Tumor volume curves from weekly MRI of Ink4a/Arf- or p53-deficient GBM-bearing mice. Two key phases were observed in both models: tumor growth in the 3 weeks preceding BLZ945 treatment (start = timepoint 0), and tumor regression in the 3 weeks post-treatment initiation.
(B) Analysis pipeline of IVM data collected from Ink4a/Arf- or p53- deficient GBM-bearing mice (CX3CR1 lineage-tracing model) that were intravitally imaged at different stages of tumor progression or therapeutic response. After segmentation of the different components of the GBM TME in each of the 30-min time-lapse experiments (step 1 and 2), brain-resident TAMs were tracked (step 3). Of note, migration of cancer cells was not observed in the total duration of the time-lapse. Data were extracted from 80 movies that were collected from 5 Ink4a/Arf-deficient GBMs and from 3 p53-deficient GBMs, measuring in total 76,036 brain-resident TAMs, and the SingleCellExperiment Bioconductor package was used to store, organize, and analyze the data (step 4). Scale bars: 100 μm.
(C) UMAP visualization of brain-resident TAM clusters from this analysis pipeline.
(D) Violin plots show the migration speed of brain-resident TAMs in each of the clusters (the cluster numbers correspond to the IDs shown in (C)).
(E) Percentage of migrating brain-resident TAMs in Ink4a/Arf- or p53-deficient GBMs at each of the stages of tumor progression (timepoints −3, −2, −1, and 0) and in response to BLZ945 (timepoints 1, 2, and 3) (Student’s t-test, ∗ indicates p < 0.05). Each dot represents one FOV. Data are represented as mean ± SD. See also Figures S3, S4, and S5, and Videos S1, S2, S3, S4, and S5.
In the multimodal imaging experiments herein, mice were imaged up to three weeks into the BLZ945 trial, the timepoint at which we observed maximal tumor regression (Figure 4A). We observed that BLZ945-treated tumors can occasionally regress toward the meninges, demonstrating the necessity of MRI to determine tumor volume. If one were to extrapolate tumor volume from the number of GBM cells under the CIW (by IVM alone), this could potentially introduce a certain amount of bias. Together, we collected data during the course of three weeks of tumor progression, and three subsequent weeks of response to treatment (up to six weeks of imaging per animal). To locate the same subpopulation of cells in an individual mouse during each of the IVM sessions, we initially acquired a tile scan of the entire tumor area under the CIW, followed by visual identification of the previously imaged fields-of-view (FOVs) (Figure 1C). The selected FOVs were intravitally imaged for 30 min (time interval = 1 to 2 min, Z-step size = 5 μm) to monitor the migration behavior of brain-resident TAMs (Videos S2, S3, S4, and S5). In total, we recorded 80 IVM movies of five Ink4a/Arf- and three p53-deficient GBM-bearing mice during tumor progression and in response to therapy, segmented the different components of the TME (the dura mater as measured by SHG imaging (Figures 3A and 3B), cancer cells, and brain-resident TAMs), tracked the migration of brain-resident TAMs, and extracted the data for quantification (Figure 4B).
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
To efficiently analyze the single-cell measurements across the different GBM models and progression/treatment timepoints, we imported data of 76,036 brain-resident TAMs into a SingleCellExperiment S4 object in R. This included features such as migration speed and distance, and metadata including MRI-based information. Next, we performed unsupervised graph-based clustering and dimensionality reduction for representation of the data (Figure 4B). Clustering analysis revealed five major clusters (Figure 4C), and when we investigated the features that distinguish these clusters, we found that cluster 3 is characterized by more migratory brain-resident TAMs (Figure 4D). We also analyzed the dataset from the FLT3 lineage-tracing model (Figures S3A and S3B). In this context, there were no specific clusters of FLT3+ cells defined by migration speed (Figure S3C). In conclusion, our single-cell imaging analysis pipeline represents a powerful tool to investigate complex and large IVM datasets. Specifically, in our dataset, it enabled the identification of different brain-resident TAM subsets, whereby their migratory ability plays a key role in defining these cellular subsets. Therefore, we next investigated the cellular migration phenotype in further detail.
Migratory ability of brain-resident TAMs differs in genetically distinct glioblastomas
In response to injury, brain-resident MG become activated and migrate to the site of damage where they phagocytose dead cells and cellular debris (Brown and Neher, 2012; Miller et al., 2019). To explore this behavior specifically in GBMs, and to discriminate cell migration from respiration-induced motion artifacts, we empirically defined the threshold for cell migration as ≥0.05 μm/s, which is equivalent to a track length of 90 μm in a 30-min time-lapse experiment (Figure S4). We observed that in response to BLZ945, brain-resident TAMs become gradually more migratory in the Ink4a/Arf-deficient model, and we observed a similar trend in p53-deficient GBMs (Figure 4E and Videos S2, S3, S4, and S5). These results are consistent with previous molecular analyses (Pyonteck et al., 2013; Yan et al., 2017), where we showed that GBM TAMs become antitumorigenic in response to different CSF-1R inhibitors by downregulating M2-like macrophage markers and enhancing phagocytosis.
Interestingly, we also revealed differences between brain-resident TAMs from Ink4a/Arf- versus p53-deficient GBMs. First, brain-resident TAMs in Ink4a/Arf-deficient GBMs are more migratory compared to their counterparts in p53-deficient GBMs, both in advanced lesions before treatment, and in response to BLZ945 (Figure 4E). Second, whereas brain-resident TAMs remain present in both tumor models following treatment initiation, consistent with our previous data where we showed that the glioma TME protects TAMs from BLZ945-mediated depletion in the Ink4a/Arf-deficient model (Pyonteck et al., 2013), brain-resident TAMs in p53-deficient GBMs were more effectively protected compared to their counterparts in Ink4a/Arf-deficient GBMs (Figure S5A). Further analyses revealed that in p53-deficient GBMs, the number of tumor cells is more substantially reduced in response to BLZ945 treatment (Figure S5B), suggesting that the abundance of antitumorigenic brain-resident macrophages in BLZ945-regressing tumors may correlate with treatment response. Future studies will be important to fully elucidate the potential translational implications of these findings.
Discussion
The TME is not a static milieu: during the course of tumor progression, and in response to therapy, the cellular composition changes considerably (Akkari et al., 2020; Magod et al., 2021). Conventional laboratory techniques allow researchers to investigate these dynamics by processing tissues at different timepoints during the course of tumor growth, or regression in response to therapy. This can be achieved by collecting and analyzing multiple time-matched specimens from different subjects; however, a large number of samples are typically required for such a time course and to account for inter-tumoral heterogeneity. Another option involves serial sampling of the same tumor at different timepoints, a method that is increasingly being incorporated into clinical trials where possible. Such experiments are currently as close as one can get to study cellular dynamics in human cancer; however, despite the invaluable information these analyses can provide, biopsies are often not taken from the same location, and the biopsy procedure itself can potentially alter the TME. It is therefore important to implement additional experimental strategies and to expand our repertoire of empirical techniques, which can complement these methods, to gain a deeper understanding of the dynamics of key biological processes, including in cancer.
IVM is such an approach, enabling cutting-edge, high-resolution cellular imaging, allowing one to visualize the cells in their natural environment, and critically to also monitor animals longitudinally. Specifically, for the multimodal imaging platform we have developed, the location of the CIW in the skull gives visual access to the brain parenchyma, but also to the meningeal layers above it. While some IVM studies of the brain removed the dura mater during the window implantation (Alieva et al., 2019), to achieve a greater imaging depth of the brain parenchyma, others left it intact (Askoxylakis et al., 2017; Ricard and Debarbieux, 2014) and could visualize it by SHG imaging (Ricard and Debarbieux, 2014). In our IVM model, the dura mater was purposely left in place to preserve the composition of the natural brain environment. Importantly, the precise source of the SHG signal was first determined by ex vivo imaging of the dura mater (signal) and the brain parenchyma (no signal).
It has been previously reported in other contexts that skull craniotomy can potentially induce local inflammation and perturb the functions of the underlying meninges and parenchyma (Coles et al., 2017). While our analyses herein did not reveal any alterations in the GBM TME following CIW implantation, in future experiments, it will be important to also assess the potential impact on normal dura homeostasis and compare it with alternative approaches such as mechanical thinning of the cranium. In another IVM study investigating the behavior and morphology of TAMs in a GBM model, image analysis starting at an imaging depth >50 μm from the pial surface was proposed as a means to differentiate the meninges and parenchyma (Chen et al., 2019). In this regard, it will also be important to consider this point for the interpretation of IVM imaging data in brain tumors.
As a consequence of these important differences in experimental design, an interesting observation we were able to make for the first time via IVM was the identification of GFP+ GBM cells within the dura. This also implies that the normal parenchymal-meningeal boundaries have been disrupted, thereby representing an additional challenge to the definition and delineation of the normal anatomical structures. Indeed, recent studies have demonstrated the presence of lymphatics in the dura mater, which drain from the brain borders to the cervical lymph nodes (Aspelund et al., 2015; Louveau et al., 2015; Petrova and Koh, 2018). Moreover, manipulation of the meningeal lymphatics in murine glioma models has been shown to represent a potent therapeutic strategy to prime T cells and induce a more effective immune response against the tumor (Hu et al., 2020; Song et al., 2020). Therefore, IVM has the unique potential to investigate the interactions between cancer cells, antigen-presenting cells, T cells, and all other TME components at the meninges-tumor interface, which could potentially be involved in regulating the adaptive immune response (or lack thereof). In order to achieve such detailed imaging of these complex cellular interactions, additional lineage-tracing models would be required as well as labeling strategies to delineate the different meningeal layers and vessels (Coles et al., 2017). These applications would also necessitate the combination of multiple fluorophores, and it will therefore be crucial to precisely define the cellular source of the different signals in future studies. Toward this end, the design and implementation of new deconvolution algorithms and spectral unmixing would facilitate the complex analysis pipeline (Kapsokalyvas et al., 2021).
IVM has been reported as a potent tool to study myeloid cell dynamics in GBM models (Chen et al., 2019; Ricard and Debarbieux, 2014; Ricard et al., 2013, 2016). These elegant studies focused on specific cell features which either varied during tumor progression (Ricard et al., 2016), or at defined endpoints in different treatment conditions (Chen et al., 2019). However, in order to achieve a full understanding of the TME dynamics over multiple longitudinal imaging sessions, it is critical to integrate all variables and interdependencies into a comprehensive analysis strategy that is capable of analyzing the large, and incredibly rich, datasets created by IVM imaging. The generation of single cell data has opened a new era of biological research, with the main focus being various omics-approaches. Imaging analysis of behavioral features and cellular morphologies allows one to similarly apply unsupervised clustering approaches to these types of data. Indeed, we found that dimensionality reduction of IVM-acquired data is a powerful strategy to define different subsets of cells. Interestingly, we identified a highly migratory cluster of CX3CR1+ brain-resident TAMs (including both MG and dural BAMs). Our current cell lineage-tracing strategy does not allow us to definitively discriminate MG from non-parenchymal macrophages. Importantly, leptomeningeal macrophages can also express CX3CR1 and are only partially replenished by peripheral immune cells (Jordao et al., 2019; Kierdorf et al., 2019). Nonetheless, we could still incorporate other parameters to more specifically delineate MG, including cellular morphology, level of Cx3cr1 expression, and distance from the SHG signal (as a measure of distance to the dura mater). Nonetheless, we cannot exclude that MG, similar to tumor cells, may also migrate into the meningeal membranes, and this will be interesting to explore further.
Future studies will also be important for a more detailed understanding of the migratory cues of TAMs following CSF-1R inhibition. These could result from cellular activation following BLZ945 treatment, and the release of chemo-attractants within the TME, but also be attributed to modifications of physical constraints (as a consequence of the substantial tumor regression) that then enable enhanced TAM migration. Furthermore, we also observed differential migratory behaviors depending on the genetic background of the tumor. Previous studies by our lab and others have shown that genetically distinct GBMs respond differentially to CSF-1R inhibition (Akkari et al., 2020; Quail et al., 2016; Rao et al., 2022), and recent reports have indicated that the genetic makeup of gliomas impacts the cellular composition of the TME (Akkari et al., 2020; Magod et al., 2021). Of particular interest is the observation that blockade of CSF-1R signaling reduced the number of TAMs in the syngeneic GL261 GBM model (Coniglio et al., 2012), whereas TAMs are protected from CSF1R inhibitor-induced apoptosis in Ink4a/Arf-deficient GBMs by secreted factors and are instead “re-educated” (Pyonteck et al., 2013). Our findings here further bolster the emerging notion that the underlying cancer cell genetics sculpt the TME, and can influence both tumor progression and response to therapy. It would thus be of interest for further studies to explore the molecular mechanisms underlying the differential migration of brain-resident TAMs in genetically distinct GBMs.
In sum, the combined use of MRI and IVM through a MRI-compatible CIW as described herein represents a powerful multimodal platform not only for investigating evolving brain malignancies but which can also be invaluable to study cellular dynamics in other pathologies of the central nervous system, such as multiple sclerosis, degenerative conditions including Alzheimer disease, and cerebral pathogen infection. Depending on the choice of the lineage-tracing mouse model and/or other labeling strategies, a plethora of cell types known to be involved in neuropathology can be visualized by IVM, including MG and other immune cells, neurons, astrocytes, oligodendrocytes, and (peri)vascular cells, while simultaneously monitoring the disease course by MRI. Future studies could even incorporate functional MRI or molecular MRI protocols to further extend the multimodal platform, similar to the imaging of iron oxide nanoparticle uptake in the TME by two-photon IVM and MRI (Karimian-Jazi et al., 2020). Lastly, we also demonstrate the critical insights gained from applying data science tools initially developed for single-cell RNA sequencing data, which facilitated the deep interrogation of large IVM datasets. These types of strategies will enable more complex and large-scale analyses in this field, for example to predict disease outcome or response to treatment.
Limitations of the study
In this study, we were able to precisely image the same area of the tumor by using specific morphological landmarks (e.g. SHG signal, or the tumor vasculature). However, as a consequence of the long time periods over which we were imaging (several weeks), we were not able to find back the exact same individual cells in successive imaging sessions. For our longitudinal experiments, we chose an imaging interval of one week, in which the tumor is either in an exponential growth phase (during tumor progression), or the tumor is regressing in response to treatment. Within this time-frame, the tumor microenvironment is substantially remodeled: tumor cells can be proliferating or dying, new immune cells are recruited from the peripheral circulation, and cells are constantly migrating. It would therefore be of interest for future studies to apply other IVM tools, such as photo-marking of individual cells, to track the fate, migration, and function of cells in the tumor microenvironment (Chtanova et al., 2014; Kedrin et al., 2008; Suan et al., 2015; Victora et al., 2010).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| BLZ945 | Novartis | N/A |
| Captisol | Ligand Pharmaceuticals | RC-0C7-K01 |
| Dental cement powder | Paladur, Kaladent | 2260240 - 100 g |
| Dental cement liquid | Paladur, Kaladent | 2260210 - 80 mL |
| Norland optical adhesive 61 | AMP Technica | 431508 |
| Deposited data | ||
| Analyzed imaging data | This paper; Mendeley Data | https://doi.org/10.17632/yt99rmbcdt.1 |
| Experimental models: Cell lines | ||
| DF1 chicken fibroblasts | ATCC | CVCL_0570 |
| Experimental models: Organisms/strains | ||
| Mouse: Nestin-Tv-a (Tg(NES-TVA)J12Ech) | Gift from Dr. E. Holland (Dai et al., 2001) | N/A |
| Mouse: Nestin-Tv-a;Ink4a/Arf−/− (Tg(NES-TVA)J12Ech; Cdkn2atm1Rdp) | Gift from Dr. E. Holland (Dai et al., 2001) | N/A |
| Mouse: Flt3:Cre; Rosa26:mTmG (Tg(Flt3-cre); Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo) | Gift from Dr. C. Forsberg (Benz et al., 2008; Boyer et al., 2011; Muzumdar et al., 2007) | N/A |
| Mouse: Cx3cr1:CreER-IRES-YFP (BL6.Cg-Cx3cr1tm2.1(cre/ERT)) | Jackson laboratories | IMSR_JAX:021160 |
| Mouse: Rosa26:lsl-TdTomato (Gt(ROSA)26Sortm14(CAG-tdTomato)) | Jackson laboratories | IMSR_JAX:007914 |
| Recombinant DNA | ||
| RCAS-PDGFB-HA | Gift from Dr. E. Holland (Becher et al., 2008; Ozawa et al., 2014) | N/A |
| RCAS-PDGFB-HA-SV40-GFP | Gift from Dr. E. Holland (Becher et al., 2008; Ozawa et al., 2014) | N/A |
| RCAS-shP53 | Gift from Dr. E. Holland (Becher et al., 2008; Ozawa et al., 2014) | N/A |
| Software and algorithms | ||
| Imaris version 9.7.2 | Bitplane | SCR_007370 |
| Code to analyze imaging data | This paper, Mendeley Data | https://doi.org/10.17632/yt99rmbcdt.1 |
| Other | ||
| Peek head bars | This paper, home-made | N/A |
| 3 mm diameter coverslips, thickness “#0” (= 0.085 mm) | Multichannel Systems | 640726 |
| 5 mm diameter coverslips, thickness “#0” (= 0.085 mm) | Multichannel Systems | 640731 |
| 3 mm biopsy punches | Integra Miltex | 33-32 |
| 3 Tesla small animal MR scanner | Bruker Biospin MRI | N/A |
| Multi-photon microscope | Leica | SCR_018852 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Johanna A. Joyce (johanna.joyce@unil.ch).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
The Nestin-Tv-a;Ink4a/Arf−/− mouse line has been described previously (Dai et al., 2001) and was generously provided by Dr. Eric Holland. The Nestin-Tv-a and Nestin-Tv-a;Ink4a/Arf−/− mouse lines were bred to a C57BL/6 background for 10 generations in the Joyce lab. Flt3:Cre; Rosa26:mTmG mice (C57BL/6 background) were kindly provided by Dr. Camilla Forsberg (UCSC) (Benz et al., 2008; Boyer et al., 2011; Muzumdar et al., 2007) and bred to Nestin-Tv-a or Nestin-Tv-a;Ink4a/Arf−/−mice (Bowman et al., 2016). Only male mice showed expression or transmittance of Cre, and as such only male mice could be used for the experiments with this line. Cx3cr1:CreER-IRES-YFP mice (C57BL/6 background; Jackson Labs) were crossed with Rosa26:lsl-TdTomato reporter mice (C57BL/6 background; Jackson Labs), and with Nestin-Tv-a or Nestin-Tv-a;Ink4a/Arf−/−mice (Bowman et al., 2016). Both male and female Cx3cr1:CreER-IRES-YFP Rosa26:lsl-TdTomato lineage-tracing mice were used for experiments (3 females and 2 males for the Ink4a/Arf-deficient GBM model, and 2 females and 1 male for the p53-deficient GBM model). For the Cx3cr1:CreER-IRES-YFP Rosa26:lsl-TdTomato lineage-tracing system, 3-4 week-old mice were injected twice, 48 h apart, intraperitoneally with 1 mg of tamoxifen citrate (Sigma-Aldrich) dissolved in sunflower seed oil (Sigma). Mice were used for intracranial injection of RCAS virus-producing DF1 cells 3 weeks after tamoxifen administration (see “Glioblastoma model” section below). All mice were bred within the University of Lausanne animal facilities, and all animal studies were approved by the Institutional Animal Care and Use Committees of the University of Lausanne and Canton Vaud, Switzerland (License numbers: VD3269 and VD3636). Littermates of the same sex were randomly assigned to the experimental groups. Mice were housed under a 12-h light/dark schedule at 22°C, in the presence of 2-5 cage mates. Standard autoclaved lab diet and water were provided.
Cells
DF1 chicken fibroblasts were obtained from the ATCC. RCAS virus vectors expressing PDGFB-HA, PDGFB-HA-SV40-GFP, or a short hairpin against mouse p53 (shP53) were kindly provided by Dr. Tatsuya Ozawa and Dr. Eric Holland (Becher et al., 2008; Ozawa et al., 2014). DF1 cells were transfected with the RCAS vectors using FuGENE 6 (Promega) according to the manufacturer’s instructions. PDGFB-HA-SV40-GFP DF1 cells were sorted for GFP-positive cells by flow cytometry to create a stable GFP-positive DF1 cell line. All cell lines were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies), and penicillin and streptomycin (Life Technologies) under standard conditions.
Glioblastoma model
4.5 to 7 week-old mice were intracranially injected as previously described (Bowman et al., 2016; Hambardzumyan et al., 2009; Pyonteck et al., 2013; Quail et al., 2016). Briefly, mice were fully anesthetized using isoflurane inhalation anesthesia (2% isoflurane/O2 mixture), and a mixture of 2% lidocaïne (Streuli Pharma) and 0.5% bupivacaine (Carbostesin; Aspen Pharma Schweiz) was applied as a local anesthetic (50 μL per mouse), and 0.3 mg/mL buprenorphine (Temgesic; Indivior Schweiz) was given subcutaneously as a systemic anesthetic (100 μL per mouse). Using a stereotactic apparatus, cells were injected into the right frontal cortex (2 mm frontal, 1.5 mm lateral from bregma, 2 mm deep). For the p53-deficient GBM model, 3 × 105 DF1 cells (a 1:1 mixture of RCAS-PDGFB-HA/PDGFB-HA-SV40-GFP DF1 cells and RCAS-shP53 DF1 cells) were injected, and for the Ink4a;Arf-deficient GBM model, mice were injected with 2 × 105 RCAS-PDGFB-HA/PDGFB-HA-SV40-GFP DF1 cells. The skin incision was sealed with Vetbond tissue adhesive (3M), and the mouse was placed on a heating pad and monitored until fully recovered from anesthesia. Finally, Bepanthen cream (Bayer) was applied on the incision site before placing the animal back in the cage, and each mouse was subsequently followed by regular monitoring.
Cranial window surgery
Mice were fully anesthetized using isoflurane inhalation anesthesia (2% isoflurane/O2 mixture) and prepared for surgery by shaving and cleaning the head with 70% ethanol pads (Fisher Healthcare) and 10% betadine pads (Purdue Products L.P.). A mixture of 2% lidocaïne and 0.5% bupivacaine was applied as a local anesthetic (50 μL per mouse), and 0.3 mg/mL buprenorphine was administered subcutaneously as a systemic anesthetic (100 μL per mouse). A longitudinal incision of the skin was made between the occiput and forehead, and the skin covering the skull was cut in a circular manner, by removing approximately 3 mm of the skin edges on each side. After cleaning and drying the skull with a cotton swab, the bone was etched mechanically with a scalpel (by making a pattern of crosses) and chemically by applying 35% phosphoric acid solution (Santa Cruz Biotechnology) with a cotton swab. The skull was then cleaned several times with Ringer’s solution (Dutscher), and the mouse was placed into a stereotactic device with a three-axis micromanipulator and an integrated heating pad (Stoelting). A 3-mm craniotomy was made using a 3-mm biopsy punch (Integra Miltex), ensuring precise fitting of the imaging window. The exposed cortical surface was kept moist with Ringer’s solution. DF1 virus-producing cells were injected into the brain parenchyma at the craniotomy site as described in the previous section. The brain surface was sealed with a 0.085-mm-thick 3-mm diameter coverslip (Multichannel Systems) that was glued to a 0.085-mm-thick 5-mm diameter coverslip (Multichannel Systems) using Norland optical adhesive 61 (AMP Technica). The outer edges of the 5-mm coverslip were used to fix the imaging window to the skull using super glue (Loctite). A small head bar was also attached using super glue, enabling the mouse’s head to remain stationary during the intravital imaging experiments. Lastly, dental acrylic glue (Kulzer) was applied to the skull surface, covering also a small rim of the 5-mm coverslip. The animal was then placed on a heating pad and monitored until it had fully recovered from anesthesia and was returned to the cage, followed by regular monitoring.
Monitoring and scoring
Upon recovery from anesthesia after the cranial window surgery, mice underwent visual observation every few hours for the first 24 h. To ensure pain relief, paracetamol (Bristol-Myers Squibb) was added to the drinking water (500 mg/250 mL) for 7 days post-operatively. Mice were monitored at least once per day for any signs of distress as well as for brain tumor symptoms, including lethargy, poor grooming, weight loss, and in rare instances macrocephaly or hemiparesis. For each sign, a severity-dependent score of 1 to 3 was given. In the event of a cumulative score of 10, or any individual score of 3, mice were immediately euthanized.
BLZ945 treatment
BLZ945 was obtained from Novartis, Switzerland, in solid state (BLZ945-AA (HCL Salt)). For in vivo experiments, BLZ945 was dissolved in 20% Captisol (w/v) (Ligand Pharmaceuticals) in H20 and stirred overnight to obtain a clear solution. Dissolved BLZ945 was administered daily at 200 mg/kg by oral gavage, as previously described (Akkari et al., 2020; Pyonteck et al., 2013; Quail et al., 2016).
Methods details
Brain slice harvesting and imaging
Mice were sacrificed by terminal anesthesia using pentobarbital (CHUV Hospital, Lausanne, Switzerland), followed by transcardial perfusion with PBS. Tumor-bearing brains were harvested and placed on ice in “slicing medium” (minimum essential media 2 mM l-glutamine (MEM GlutaMAX, Life Technologies), 4.5 mg/mL glucose (Life Technologies), 100 U/mL penicillin-streptomicin (P/S, Life Technologies)). The brain was then fixed on the specimen plate of a vibratome (Leica) with super glue (Loctite). The brain was held in place with 2% agar cubes to additionally support the tissue while slicing, and submerged into “slicing medium” in the buffer tray. The vibratome was set at a moving speed of 0.5 mm/s, a vibration amplitude of 1 μm, and 425-μm-thick coronal slices were cut sequentially and placed into “culturing medium” (MEM GlutaMAX, 4.5 mg/mL glucose, 100 U/mL P/S, 25% HBSS (Life Technologies), 25% heat-inactivated normal horse serum; re-supplement: 1X GlutaMAX (Life Technologies), 1X non-essential amino acids (Life Technologies), and 1:50 1M HEPES (Life Technologies)) on ice. 3 to 4 slices per mouse were placed on a microscope slide (Fisher Scientific) and enclosed by a layer of 732 multi-purpose sealant (Dow) to create a small bucket which was then filled with a small amount of “culturing medium”, and finally enclosed by a coverslip (Menzel-Gläser). Images were acquired using an upright Leica TCS SP8 DIVE multi-photon microscope (Mannheim, Germany) equipped with a 16× (HC FLUOTAR L N.A. 0.6 FWD 2.5 mm) multi-immersion objective with the same imaging parameters as for IVM (see “Intravital imaging” section below).
Flow cytometry
Mice were sacrificed by terminal anesthesia using pentobarbital, followed by transcardial perfusion with PBS (CHUV Hospital Lausanne, Switzerland). The brain tumor was macrodissected and dissociated using the Brain Tumor Dissociation Kit (Miltenyi). The dissociated tissue was filtered through a 40 μm mesh filter and then subjected to myelin removal using the Myelin Removal Beads Kit (Miltenyi). The resulting myelin-free single cell suspension underwent red blood cell lysis for 10 min on ice using 10× red blood cell lysis buffer (Biolegend). The single cell suspension was stained with the Zombie-near-infrared fixable viability kit (Biolegend) for 20 min at room temperature (RT), washed with FACS buffer (2 mM EDTA (Life Technologies) and 0.5% BSA (Jackson ImmunoResearch) in PBS), and then FC-blocked (BD Biosciences) for 30 min on ice. After washing with FACS buffer, cells were incubated with directly-conjugated antibodies for 30 min on ice (Table S1). Stained samples were then washed 3 times with FACS buffer and acquired on a BD Fortessa at the Flow Cytometry Core Facility of the University of Lausanne.
Immunofluorescence staining and imaging of the dura mater
Mice were sacrificed by terminal anesthesia using pentobarbital, followed by transcardial perfusion with PBS. The skull was collected and fixed overnight in periodate-lysine-paraformaldehyde (PLP) buffer at 4°C. The dura mater was then manually removed from the skull bone, placed in a 24-well plate and washed for 10 min in 500 μL of PBS on a shaker at RT. The dura mater was next permeabilized and blocked for 2 h with TritonX-100 0.5% (AppliChem) and 5% normal donkey serum (NDS) (EMD Millipore) in PBS on a shaker at RT. After removal of the permeabilization/blocking buffer, the dura mater was incubated for 20 h in 300 μL of primary antibody mix (Table S1) diluted in 5% NDS/PBS on a shaker at 4°C. After washing 3 times (10 min each) in 500 μL PBS on a shaker at RT, the sample was incubated for 2 h with 300 μL of secondary antibody mix (Table S1) diluted in 5% NDS/PBS on a shaker at RT (protected from the light). After washing 3 times (10 min each) in 500 μL PBS on a shaker at RT, the sample was incubated for 15 min in 300 μL DAPI (Life Technologies) (1:5000 in PBS) on the shaker at RT. The dura mater was placed on a glass slide (Fisher Scientific), mounted with Dako fluorescence mounting medium (Agilent), and covered with a coverslip. Images were acquired using an upright Leica TCS SP8 DIVE multi-photon microscope (Mannheim, Germany) equipped with a 16× (HC FLUOTAR L N.A. 0.6 FWD 2.5 mm) multi-immersion objective corrected for imaging with H2O. The imaging parameters were similar to the parameters used for IVM (to visualize SHG signal, GFP and tdTomato - see “Intravital imaging” section below), and the 633 nm laser was used to visualize the collagen type I antibody that was labeled with AF647.
Magnetic resonance imaging (MRI)
MR imaging was performed using a 3 Tesla small animal MR scanner (Bruker BioSpin MRI, Ettlingen, Germany) with an 82-mm volume coil as transmitter combined with a 2 × 2 mouse brain phased array surface coil for signal reception. The mouse was fully anaesthetized with 1–2% isoflurane/oxygen inhalation and placed on the imaging bed with the head held in place below the surface coil, while monitoring respiratory rate and temperature. Data acquisition was performed using the Paravision 360 v2.0 software (Bruker BioSpin MRI, Ettlingen, Germany). A 3-slice localizer was performed to assess the mouse head position. 2D turbo rapid acquisition relaxation enhancement (Turbo-RARE) T2-weighted acquisition was performed with the following pulse sequence parameters: TR = 3000 ms, TE = 75 ms, NA = 6, number of slices 10, slice thickness (ST) = 0.7 mm, FOV = 20 × 20 mm2, pixel size 0.156 × 0.156 mm2, (ETL = 12, Tacq = 3 min) with images being acquired in axial planes. After imaging, the mouse was returned to the cage and monitored until it regained consciousness. Volumetric analysis of the GBMs was performed on MRI DICOM files using the MIPAV software (National Institutes of Health, USA).
Intravital 2-photon imaging
Animals were sedated using isoflurane inhalation anesthesia (3% isoflurane/O2 mixture), and mice that were used for vascular imaging were intravenously injected with Pacific Blue-labeled 500 kDa dextran (both Thermo Fisher Scientific, and prepared according to the manufacturers’ instructions). Mice were fixed with their head-bar to a custom-designed mouse holder (CHUV Hospital workshop, Lausanne, Switzerland), and the concentration of isoflurane was lowered to 0.8–1.2% to keep the mice lightly sedated, characterized by a constant and non-forced breathing pattern. This level of isoflurane is optimal for imaging sessions up to several hours, and it minimizes motion-induced image deformation caused by irregular breathing. The mouse was placed in the climate chamber surrounding the microscope, keeping the whole stage of the microscope and the objectives at 34°C. Imaging was performed on an upright Leica TCS SP8 DIVE multi-photon microscope (Mannheim, Germany) with an InSight X3 tunable laser (Spectra Physics) and two non-descanned hybrid 4Tune detectors. SHG imaging was performed using a wavelength of 810 nm for excitation, and 380–420 nm for detection. GFP and tdTomato were excited with a wavelength of 960 nm, and detected between 500-540 nm and 570–670 nm respectively. Pacific Blue-labeled dextran was excited using a wavelength of 810 nm, and detected between 435 and 480 nm. All images were acquired with a 16× multi-immersion objective (HC FLUOTAR L N.A. 0.6 FWD 2.5 mm). The objective was corrected for imaging with Immersol W 2010 (Carl Zeiss) and a coverslip thickness of 0.17 mm. For each mouse, a tile scan of the whole CIW was taken. Afterwards, two individual time-lapses of 30 min each were acquired. The exact location of previously acquired fields-of-view was retrieved by visual identification of clear landmarks such as SHG imaging patterns and vasculature, as well as by using previously saved coordinates of the stage of the microscope.
Imaris image analysis
High-resolution three-dimensional time-lapse images were imported into Imaris version 9.7.2 (Bitplane). Before cell/structure classification and segmentation, images were corrected for bleed-through using the channel arithmetic function. MG, tumor cells, and ECM (as measured by SHG imaging) were classified and segmented using the surface creation tool. Features of individual surfaces were exported as.csv files for each individual time-lapse.
Data import into R
Individual .csv files per imaging session were loaded into R version 4.0.2, using R Studio version 1.4.1103. For each imaging session, the following features were extracted per cell: shortest distance of MG to SHG, shortest distance of MG to tumor cells, displacement length, track duration, ellipticity oblate, ellipticity prolate, intensity of GFP, intensity of tdTomato, track length, number of surfaces, track position, track speed, volume, sphericity, and track straightness. For the features “shortest distance of MG to SHG” and “shortest distance of MG to tumor cells”, a measurement was made for each imaged timepoint during that imaging session. From these individual values, a mean value was calculated of all timepoints using the dplyr package version 1.0.3. All other features were exported from Imaris as mean values of all timepoints. Thus, a single value was obtained for each feature and for each cell per imaging session. All cells from all imaging sessions of all mice were then agglomerated, with annotation for track position (x, y and z), number of surfaces, track duration, imaging date, mouse ID, imaging position, surface, tracing model, tumor model, tumor volume as measured by MRI, BLZ945 start date, the day relative to BLZ945 start date, uniform timepoint, and comments in the metadata. This resulted in a dataset of 76,036 MG with 12 features, and 16 metadata categories.
Scaling, dimensionality reduction and clustering
Data were loaded into an S4 object of the SingleCellExperiment class, using the SingleCellExperiment package version 1.10.1, with the data added to the counts assay and metadata loaded in colData. For dimensionality reduction and clustering, data were scaled and centered. For any missing timepoints (some imaging timepoints were missing data for shortest distance to SHG or tumor cells), NAs were replaced with the mean for that feature of all other cells. Scaled data were added to the normcounts assay of the SingleCellExperiment class object. PCA was performed using the prcomp function, and UMAP dimensionality reduction was calculated using the RunUMAP function from the Seurat package version 4.0.1, both using default settings. Both dimensionality reductions were added to the SingleCellExperiment class object. Clustering was performed by consecutively running the FindNeighbors (k.param set to 30) and FindClusters (resolution set to 0.19) functions from the Seurat package version 4.0.1 on the scaled data. Clustering classifications were added as ‘clust’ to the colData of the SingleCellExperiment class object.
Data visualization
Data were plotted using GraphPad Prism version 9.0.1 or in R using the ggplot2 package, version 3.3.3. Labels were plotted using the ggrepel package, version 0.9.1, and color schemes were used from viridis package, version 0.6.0. For visualization, the data in the counts assay, not the scaled data in the normcounts assay, were plotted.
Quantification and statistical analysis
GraphPad Prism version 9.0.1 or R Studio was used for all data analysis. Parametric data were analyzed by a two-tailed Student’s t test. Non-parametric data were analyzed by a Mann-Whitney test on ranks. p < 0.05 was considered as statistically significant in all cases. The specific statistical test used is indicated for each experiment in the figure legend.
Acknowledgments
We thank members of the Joyce lab for insightful discussion, Lucie Tillard and Dr. Marta J.C Jordão for technical support, and Novartis Switzerland for providing BLZ945 free of charge. We are grateful to the In Vivo Imaging Facility at the University of Lausanne for assistance and advice, in particular Dr. A. Benechet. We thank Dr. E. Holland for generously providing the RCAS vectors and Nestin-Tva mice, and Dr. C. Forsberg for the Flt3:Cre Rosa26:mTmG mice. Research in the Joyce lab for this project was provided in part by the Charlie Teo Foundation, Swiss Cancer Research Foundation (KFS-3390-08-2016; KFS-5280-02-2021), the Ludwig Institute for Cancer Research, and the University of Lausanne. AZ was supported in part by a fellowship from the Human Frontier Science Program (LT000240/2017-L) and a Veni fellowship from the Netherlands Organisation for Scientific Research (09150161910076).
Author contributions
A.Z., D.C., J.K., and J.A.J. conceived the study, designed experiments, and interpreted data. A.Z., D.C., and J.A.J. wrote the manuscript. A.Z., D.C., and J.K. performed the experiments. A.Z. and D.C. performed the experimental analyses. L.v.G. performed the computational analyses. J.A.J. supervised the research. All authors edited or commented on the manuscript.
Declaration of interests
The authors declare no competing interests. J.K. currently works at Lunaphore, Switzerland, and L.v.G at Genmab, The Netherlands; J.A.J. has received honoraria for speaking at research symposia organized by Bristol Meyers Squibb and Glenmark Pharmaceuticals, and currently serves on the scientific advisory board of Pionyr Immunotherapeutics.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104570.
Supplemental information
Data and code availability
-
•
Imaging data analyzed by and exported from Imaris have been deposited at Mendeley data. The DOI is listed in the Key resources table. Two-photon microscopy images and time-lapses reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at Mendeley data and is publicly available as of the date of publication. DOIs are listed in the Key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akkari L., Bowman R.L., Tessier J., Klemm F., Handgraaf S.M., de Groot M., Quail D.F., Tillard L., Gadiot J., Huse J.T., et al. Dynamic changes in glioma macrophage populations after radiotherapy reveal CSF-1R inhibition as a strategy to overcome resistance. Sci. Transl. Med. 2020;12:eaaw7843. doi: 10.1126/scitranslmed.aaw7843. [DOI] [PubMed] [Google Scholar]
- Alieva M., Leidgens V., Riemenschneider M.J., Klein C.A., Hau P., van Rheenen J. Intravital imaging of glioma border morphology reveals distinctive cellular dynamics and contribution to tumor cell invasion. Sci. Rep. 2019;9:2054. doi: 10.1038/s41598-019-38625-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis C.D., Ferraro G.B., Jain R.K. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer. 2020;20:26–41. doi: 10.1038/s41568-019-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askoxylakis V., Badeaux M., Roberge S., Batista A., Kirkpatrick N., Snuderl M., Amoozgar Z., Seano G., Ferraro G.B., Chatterjee S., et al. A cerebellar window for intravital imaging of normal and disease states in mice. Nat. Protoc. 2017;12:2251–2262. doi: 10.1038/nprot.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspelund A., Antila S., Proulx S.T., Karlsen T.V., Karaman S., Detmar M., Wiig H., Alitalo K. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 2015;212:991–999. doi: 10.1084/jem.20142290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher O.J., Hambardzumyan D., Fomchenko E.I., Momota H., Mainwaring L., Bleau A.M., Katz A.M., Edgar M., Kenney A.M., Cordon-Cardo C., et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res. 2008;68:2241–2249. doi: 10.1158/0008-5472.CAN-07-6350. [DOI] [PubMed] [Google Scholar]
- Bejarano L., Jordāo M.J., Joyce J.A. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11:933–959. doi: 10.1158/2159-8290.CD-20-1808. [DOI] [PubMed] [Google Scholar]
- Benz C., Martins V.C., Radtke F., Bleul C.C. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J. Exp. Med. 2008;205:1187–1199. doi: 10.1084/jem.20072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman R.L., Klemm F., Akkari L., Pyonteck S.M., Sevenich L., Quail D.F., Dhara S., Simpson K., Gardner E.E., Iacobuzio-Donahue C.A., et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17:2445–2459. doi: 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer S.W., Schroeder A.V., Smith-Berdan S., Forsberg E.C. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9:64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.C., Neher J.J. Eaten alive! Cell death by primary phagocytosis: 'phagoptosis'. Trends Biochem. Sci. 2012;37:325–332. doi: 10.1016/j.tibs.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Chen Z., Ross J.L., Hambardzumyan D. Intravital 2-photon imaging reveals distinct morphology and infiltrative properties of glioblastoma-associated macrophages. Proc. Natl. Acad. Sci. U S A. 2019;116:14254–14259. doi: 10.1073/pnas.1902366116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T., Hampton H.R., Waterhouse L.A., Wood K., Tomura M., Miwa Y., Mackay C.R., Brink R., Phan T.G. Real-time interactive two-photon photoconversion of recirculating lymphocytes for discontinuous cell tracking in live adult mice. J. Biophot. 2014;7:425–433. doi: 10.1002/jbio.201200175. [DOI] [PubMed] [Google Scholar]
- Coles J.A., Myburgh E., Brewer J.M., McMenamin P.G. Where are we? The anatomy of the murine cortical meninges revisited for intravital imaging, immunology, and clearance of waste from the brain. Prog. Neurobiol. 2017;156:107–148. doi: 10.1016/j.pneurobio.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Coniglio S.J., Eugenin E., Dobrenis K., Stanley E.R., West B.L., Symons M.H., Segall J.E. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol. Med. 2012;18:519–527. doi: 10.2119/molmed.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C., Celestino J.C., Okada Y., Louis D.N., Fuller G.N., Holland E.C. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebel E., Kapolou K., Unger S., Nunez N.G., Utz S., Rushing E.J., Regli L., Weller M., Greter M., Tugues S., et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell. 2020;181:1626–1642.e20. doi: 10.1016/j.cell.2020.04.055. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D., Amankulor N.M., Helmy K.Y., Becher O.J., Holland E.C. Modeling adult gliomas using RCAS/t-va technology. Transl. Oncol. 2009;2:89–IN6. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Deng Q., Ma L., Li Q., Chen Y., Liao Y., Zhou F., Zhang C., Shao L., Feng J., et al. Meningeal lymphatic vessels regulate brain tumor drainage and immunity. Cell Res. 2020;30:229–243. doi: 10.1038/s41422-020-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordão M.J.C., Sankowski R., Brendecke S.M., Sagar. Locatelli G., Tai Y.H., Tay T.L., Schramm E., Armbruster S., Hagemeyer N., Hagemeyer N., et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363:eaat7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- Kapsokalyvas D., Rosas R., Janssen R.W.A., Vanoevelen J.M., Nabben M., Strauch M., Merhof D., van Zandvoort M.A.M.J. Multiview deconvolution approximation multiphoton microscopy of tissues and zebrafish larvae. Sci. Rep. 2021;11:10160. doi: 10.1038/s41598-021-89566-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimian-Jazi K., Münch P., Alexander A., Fischer M., Pfleiderer K., Piechutta M., Karreman M.A., Solecki G.M., Berghoff A.S., Friedrich M., et al. Monitoring innate immune cell dynamics in the glioma microenvironment by magnetic resonance imaging and multiphoton microscopy (MR-MPM) Theranostics. 2020;10:1873–1883. doi: 10.7150/thno.38659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D., Gligorijevic B., Wyckoff J., Verkhusha V.V., Condeelis J., Segall J.E., van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat. Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K., Masuda T., Jordão M.J.C., Prinz M. Macrophages at CNS interfaces: ontogeny and function in health and disease. Nat. Rev. Neurosci. 2019;20:547–562. doi: 10.1038/s41583-019-0201-x. [DOI] [PubMed] [Google Scholar]
- Klemm F., Joyce J.A. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015;25:198–213. doi: 10.1016/j.tcb.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm F., Maas R.R., Bowman R.L., Kornete M., Soukup K., Nassiri S., Brouland J.P., Iacobuzio-Donahue C.A., Brennan C., Tabar V., et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181:1643–1660.e17. doi: 10.1016/j.cell.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komohara Y., Ohnishi K., Kuratsu J., Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008;216:15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- Lathia J.D., Gallagher J., Myers J.T., Li M., Vasanji A., McLendon R.E., Hjelmeland A.B., Huang A.Y., Rich J.N. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS One. 2011;6:e24807. doi: 10.1371/journal.pone.0024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau A., Smirnov I., Keyes T.J., Eccles J.D., Rouhani S.J., Peske J.D., Derecki N.C., Castle D., Mandell J.W., Lee K.S., et al. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523:337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magod P., Mastandrea I., Rousso-Noori L., Agemy L., Shapira G., Shomron N., Friedmann-Morvinski D. Exploring the longitudinal glioma microenvironment landscape uncovers reprogrammed pro-tumorigenic neutrophils in the bone marrow. Cell Rep. 2021;36:109480. doi: 10.1016/j.celrep.2021.109480. [DOI] [PubMed] [Google Scholar]
- Miller E.B., Zhang P., Ching K., Pugh E.N., Jr., Burns M.E. In vivo imaging reveals transient microglia recruitment and functional recovery of photoreceptor signaling after injury. Proc. Natl. Acad. Sci. U. S. A. 2019;116:16603–16612. doi: 10.1073/pnas.1903336116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Ochocka N., Segit P., Walentynowicz K.A., Wojnicki K., Cyranowski S., Swatler J., Mieczkowski J., Kaminska B. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat. Commun. 2021;12:1151. doi: 10.1038/s41467-021-21407-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M., Jung E., Sahm F., Solecki G., Venkataramani V., Blaes J., Weil S., Horstmann H., Wiestler B., Syed M., et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528:93–98. doi: 10.1038/nature16071. [DOI] [PubMed] [Google Scholar]
- Ott M., Prins R.M., Heimberger A.B. The immune landscape of common CNS malignancies: implications for immunotherapy. Nat. Rev. Clin. Oncol. 2021;18:729–744. doi: 10.1038/s41571-021-00518-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T., Riester M., Cheng Y.K., Huse J.T., Squatrito M., Helmy K., Charles N., Michor F., Holland E.C. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288–300. doi: 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst C.N., Yang G., Ninan I., Savas J.N., Yates J.R., 3rd, Lafaille J.J., Hempstead B.L., Littman D.R., Gan W.B. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova T.V., Koh G.Y. Organ-specific lymphatic vasculature: from development to pathophysiology. J. Exp. Med. 2018;215:35–49. doi: 10.1084/jem.20171868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protasoni M., Sangiorgi S., Cividini A., Culuvaris G.T., Tomei G., Dell'Orbo C., Raspanti M., Balbi S., Reguzzoni M. The collagenic architecture of human dura mater. J. Neurosurg. 2011;114:1723–1730. doi: 10.3171/2010.12.JNS101732. [DOI] [PubMed] [Google Scholar]
- Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F., Olson O.C., Quick M.L., Huse J.T., Teijeiro V., et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Bowman R.L., Akkari L., Quick M.L., Schuhmacher A.J., Huse J.T., Holland E.C., Sutton J.C., Joyce J.A. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352:aad3018. doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31:326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R., Han R., Ogurek S., Xue C., Wu L.M., Zhang L., Zhang L., Hu J., Phoenix T.N., Waggoner S.N., Lu Q.R. Glioblastoma genetic drivers dictate the function of tumor-associated macrophages/microglia and responses to CSF1R inhibition. Neuro Oncol. 2022;24:584–597. doi: 10.1093/neuonc/noab228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard C., Debarbieux F.C. Six-color intravital two-photon imaging of brain tumors and their dynamic microenvironment. Front. Cell. Neurosci. 2014;8:57. doi: 10.3389/fncel.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard C., Stanchi F., Rodriguez T., Amoureux M.C., Rougon G., Debarbieux F. Dynamic quantitative intravital imaging of glioblastoma progression reveals a lack of correlation between tumor growth and blood vessel density. PLoS One. 2013;8:e72655. doi: 10.1371/journal.pone.0072655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard C., Tchoghandjian A., Luche H., Grenot P., Figarella-Branger D., Rougon G., Malissen M., Debarbieux F. Phenotypic dynamics of microglial and monocyte-derived cells in glioblastoma-bearing mice. Sci. Rep. 2016;6:1. doi: 10.1038/srep26381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits M. MRI biomarkers in neuro-oncology. Nat. Rev. Neurol. 2021;17:486–500. doi: 10.1038/s41582-021-00510-y. [DOI] [PubMed] [Google Scholar]
- Song E., Mao T., Dong H., Boisserand L.S.B., Antila S., Bosenberg M., Alitalo K., Thomas J.-L., Iwasaki A. VEGF-C-driven lymphatic drainage enables immunosurveillance of brain tumours. Nature. 2020;2020 577:7792. doi: 10.1038/s41586-019-1912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Suan D., Nguyen A., Moran I., Bourne K., Hermes J.R., Arshi M., Hampton H.R., Tomura M., Miwa Y., Kelleher A.D., et al. T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity. 2015;42:704–718. doi: 10.1016/j.immuni.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk S.J.E., van Rheenen J. From good to bad: intravital imaging of the hijack of physiological processes by cancer cells. Develop. Biol. 2017;428:328–337. doi: 10.1016/j.ydbio.2017.04.015. [DOI] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., Nussenzweig M.C. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenstein M.D., de Visser K.E. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48:399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Yan D., Kowal J., Akkari L., Schuhmacher A.J., Huse J.T., West B.L., Joyce J.A. Inhibition of colony stimulating factor-1 receptor abrogates microenvironment-mediated therapeutic resistance in gliomas. Oncogene. 2017;36:6049–6058. doi: 10.1038/onc.2017.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peripheral FLT3+ cells are shown in yellow and tdTomatoHIGH cells in violet. The total duration of the time-lapse is approximately 30 min with an interval of 1 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Brain-resident TAMs are shown in red and tumor cells in green, and the total duration of each time-lapse is approximately 30 min with an interval of 1–2 min between each frame.
Data Availability Statement
-
•
Imaging data analyzed by and exported from Imaris have been deposited at Mendeley data. The DOI is listed in the Key resources table. Two-photon microscopy images and time-lapses reported in this paper will be shared by the lead contact upon request.
-
•
All original code has been deposited at Mendeley data and is publicly available as of the date of publication. DOIs are listed in the Key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.