Figure 5.
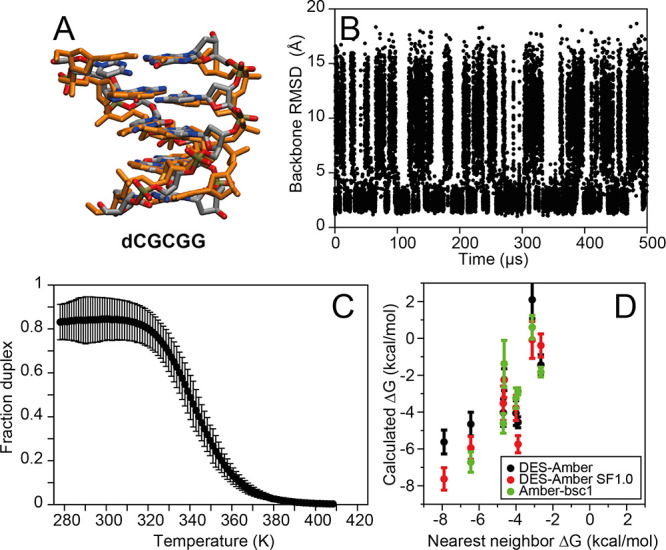
Simulated tempering simulations of short B-DNA fragments. (A) Snapshot from the d(CGCGG) simulation superimposed on a canonical B-DNA reference structure. (B) Backbone RMSD from a canonical B-DNA structure in the simulation of d(CGCGG) performed with DES-Amber. (C) Fraction of B-DNA duplex frames as a function of temperature. (D) Calculated melting free energy at 300 K for short B-DNA duplexes from simulations performed with DES-Amber, DES-Amber SF1.0, and Amber-bsc1 compared to the melting free energy predicted using a nearest-neighbor model.
