Figure 7.
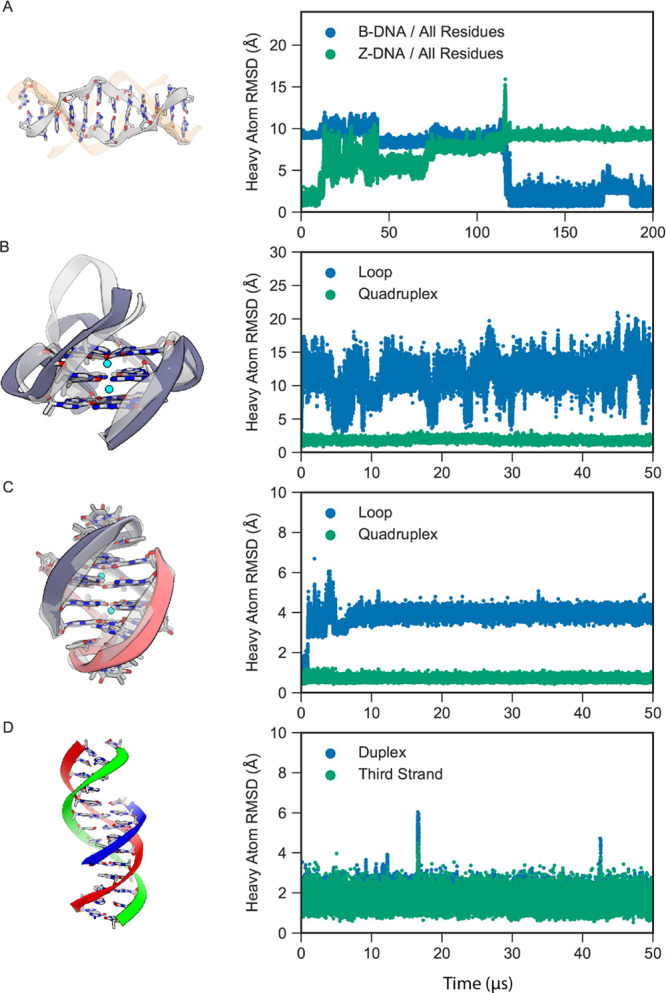
MD simulations of other forms of DNA with DES-Amber. (A) A Z-DNA dodecamer (gray) in 0.75 M KCl spontaneously transitioned to B-DNA (orange). The heavy-atom RMSDs with respect to the initial Z-DNA structure (green) and final B-DNA structure (blue) are shown on the right. (B) The CEB25 human minisatellite locus G-quadruplex, with the initial structure shown in light gray and the final structure shown in dark gray. The heavy-atom RMSDs of the loop (blue) and quadruplex (green) segments (in the reference alignment to the quadruplex core) are shown on the right. (C) The Oxytricha nova telomeric DNA G-quadruplex, with the initial structure shown in light gray and the final structure shown in dark gray and red. The heavy-atom RMSDs of the loop (blue) and quadruplex (green) segments (in the reference alignment to the quadruplex core) are shown on the right. (D) Triple-stranded DNA, with the duplex shown in red and green and the third strand in blue. The heavy-atom RMSDs of the duplex (blue) and third strand (green) (in the reference alignment of the duplex) are shown on the right.
