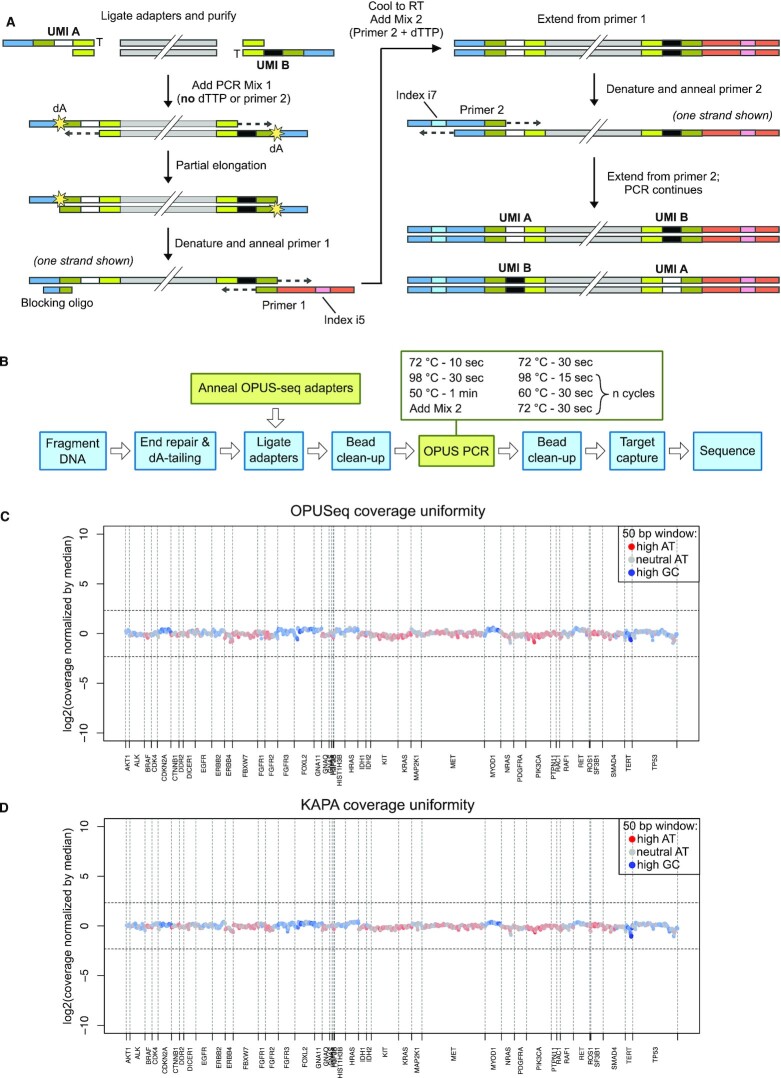
Figure 1.
OPUSeq workflow and performance in target capture. (A) Fragmented, end-repaired and dA-tailed DNA is ligated to OPUSeq adapters. Each ligated adapter carries a different single-stranded UMI. The ligated DNA is mixed with PCR mix 1 and subjected to partial elongation, which copies the UMI over to the opposite strand. The elongation stops at the first dA due to lack of dTTP in mix 1. Next, DNA is denatured and primer 1 and blocking oligo are annealed. The reaction is brought to room temperature and primer 2 and dTTP are added (mix 2). Another elongation step ensures full extension of primer 1. Finally, the desired number of standard PCR cycles are performed. (B) Schematic showing how OPUSeq is integrated into a standard PCR-based library preparation approach (KAPA). (C) Coverage uniformity for an OPUSeq library made from 5 ng gDNA and captured on a 56-kb cancer-related gene panel with a 22-kb target region. Coverage was calculated in bins of 50 bp, normalized by cross-panel median, log2 transformed and plotted per bin. Color indicates GC content of each bin. X-axis is labeled with the names of the targeted genes. (D) Coverage uniformity as in panel (C), but for a library generated from 5 ng gDNA following the standard KAPA HyperPlus protocol.