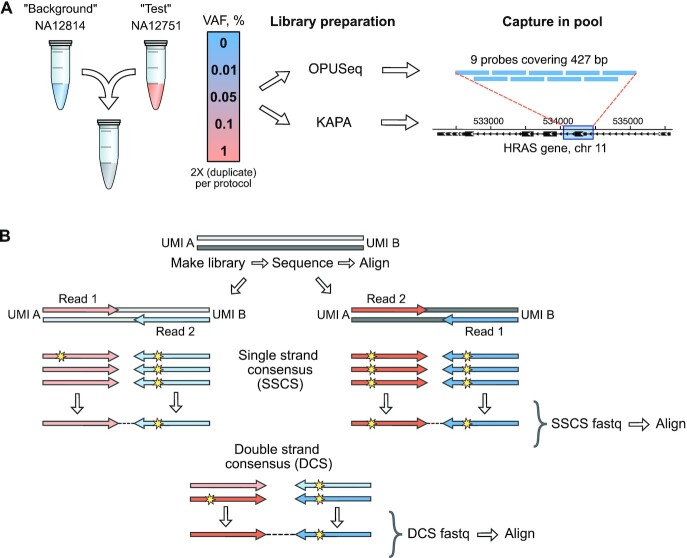
Figure 2.
Experimental design and computational workflow for low VAF detection. (A) Validation experiment to demonstrate low VAF detection with OPUSeq. Two genotypes were assigned as ‘background’ and ‘test’. They were mixed at 0–1% VAF of ‘test’ in ‘background’ in duplicate. Each gDNA mix was split into two halves: one half was subjected to OPUSeq and the other to standard KAPA protocol. The resulting libraries were captured on probes covering 427 bp of the HRAS gene on chromosome 11. (B) Overview of the computational workflow for OPUSeq data analysis. After library PCR, each original strand of a DNA duplex gives rise to its own PCR product, and each of these results in two read families (reads 1 and 2). After aligning reads to the human genome, tag families are formed by grouping reads with the same genomic coordinates, orientation and UMI. The reads within each tag family are compared to form SSCS. Only bases that are present in over two-thirds of reads are kept, which excludes errors. Finally, SSCS from opposing strands are compared to form DCS.