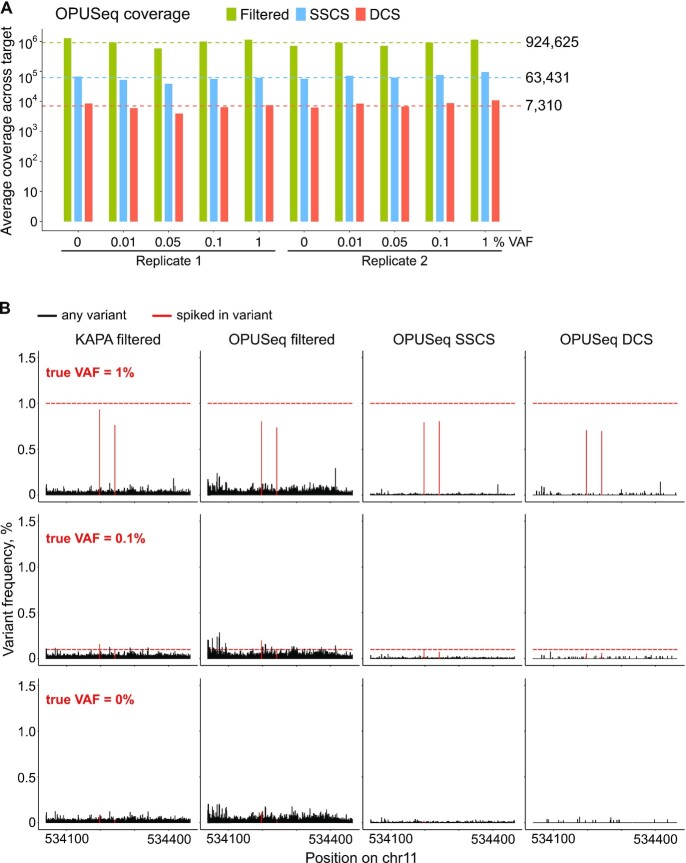
Figure 3.
OPUSeq coupled with a fragmentase-based approach helps to remove most, but not all, spurious variants. (A) Log10-transformed average coverage across target by filtered, SSCS and DCS aligned reads in the 10 sequenced OPUSeq samples obtained using the fragmentase-based protocol. The dashed lines and numbers mark the cross-sample averages. (B) Variant frequencies (%) of all detected variants per position in the target region in KAPA filtered, OPUSeq filtered, OPUSeq SSCS and DCS reads (fragmentase protocol). The spiked-in variants are labeled in red. The red dashed lines mark the expected variant frequency of the spiked-in variants. The samples shown are from replicate 2.