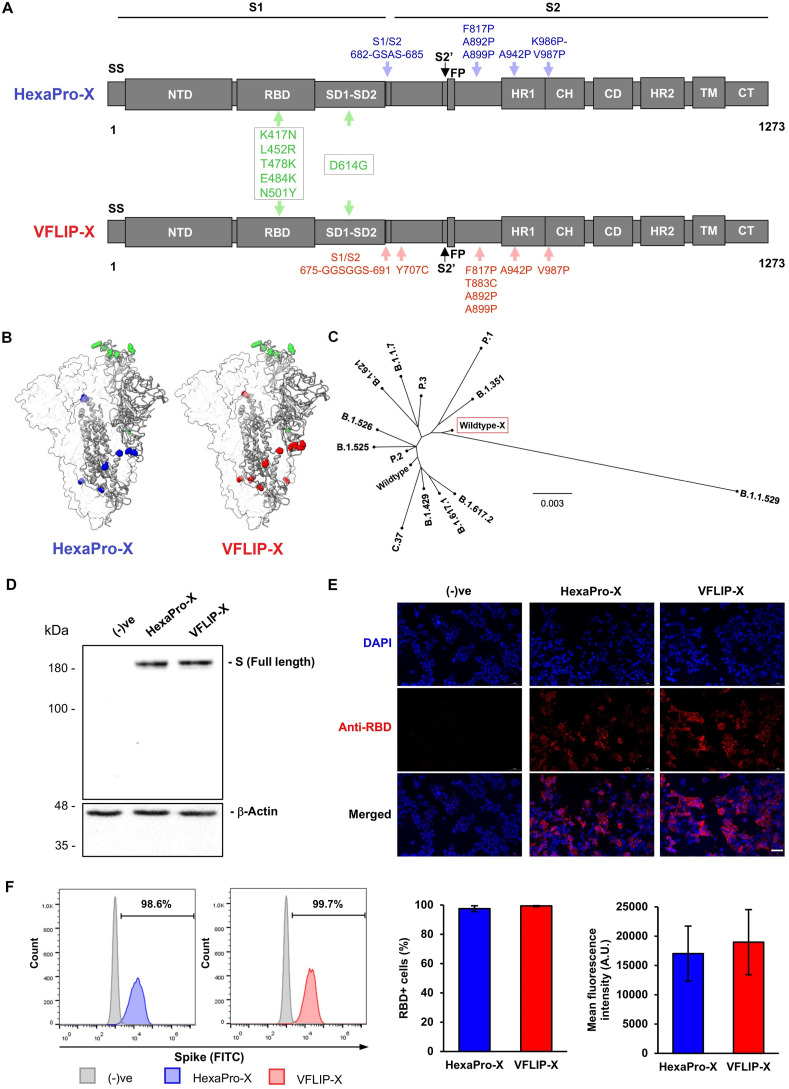
Fig. 1.
Design strategy and expression of SARS-CoV-2 spike proteins harboring six rationally substituted amino acids. (A) Schematic representation of HexaPro-X and VFLIP-X showing the S1 and S2 subunits. Amino acid positions described in the original HexaPro (Hsieh et al., 2020) and VFLIP (Olmedillas et al., 2021) spikes are shown in blue and red colors, respectively. The positions of six rationally substituted amino acids (K417N, L452R, T478K, E484K, N501Y and D614G) are indicated by green color. (B) Molecular models of HexaPro-X and VFLIP-X spike trimers. The RBD-up protomer is shown in ribbons colored corresponding to (A). The structures were prepared in SWISS-MODEL. (C) An unrooted phylogenetic tree comparing amino acid sequences derived from the wildtype spike containing six rationally substituted amino acids (wildtype-X) and spike sequences derived from SARS-CoV-2 VOCs and VOIs. (D) Western blot analysis of full-length SARS-CoV-2 spike expression in HEK293T cells transfected with circRNAs encoding HexaPro-X and VFLIP-X. The proteins were visualized by an anti-RBD antibody. (E) Immunofluorescence analysis of HexaPro-X and VFLIP-X in HEK293T cells immunostained by an anti-RBD antibody. Scale bar, 50 μm. (F) Flow cytometry analysis of HEK293T cells expressing HexaPro-X and VFLIP-X spikes upon delivery by circRNA-LNP. The proteins were visualized by an anti-RBD antibody. Percentage of RBD-positive cells and mean fluorescence intensity from biological duplicates are shown. The error bars indicate the ±SD.