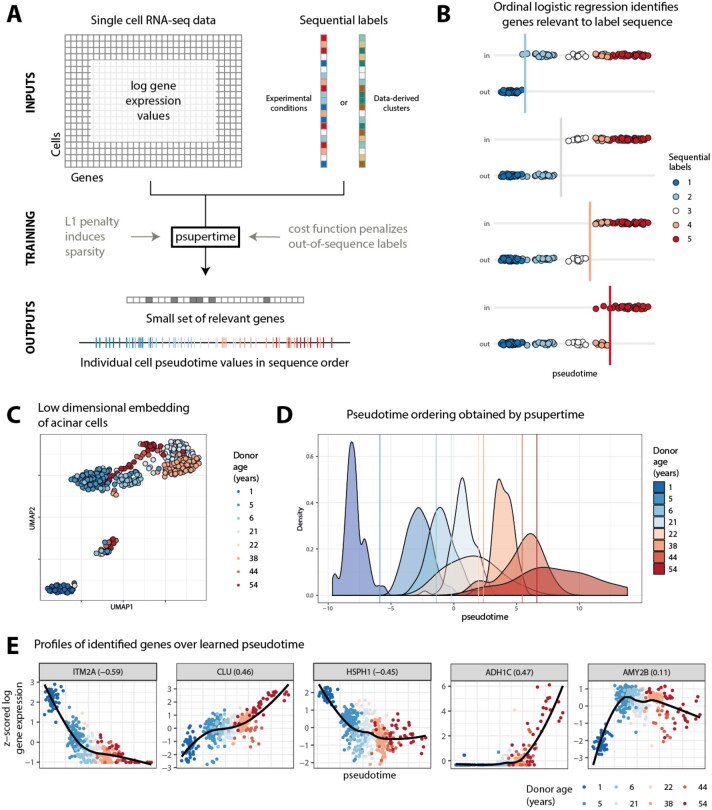
Fig. 1.
(A) Inputs to psupertime are single-cell RNA-seq data, where the cells have sequential labels associated with them. psupertime then identifies a sparse set of ordering coefficients for the genes. Multiplying the gene expression values by this vector of coefficients gives pseudotime values for each cell, which place the labels approximately in sequence. (B) Cartoon of statistical model used by psupertime, including thresholds between labels. Where there is a sequence of K condition labels, psupertime learns K−1 simultaneous (i.e. sharing coefficients) logistic regressions, each seeking to separate labels (out) from (in). (C) Dimensionality reduction of 411 human acinar cell data with ages ranging from 1 to 54 (Enge et al., 2017). Representations in two dimensions via non-linear dimensionality reduction technique UMAP. Colours indicate donor age. (D) Distributions of donor ages for acinar cells over the pseudotime learned psupertime. Vertical lines indicate thresholds learned by psupertime distinguishing between earlier and later sets of labels; colour corresponds to the next later label. (E) Expression values of selected genes (five with largest absolute coefficients; see Supplementary Fig. S2 for 20 largest). The x-axis is psupertime value learned for each cell; y-axis is z-scored gene expression values. Gene labels also show the Kendall’s τ correlation between sequential labels (treated as a sequence of integers ) and gene expression