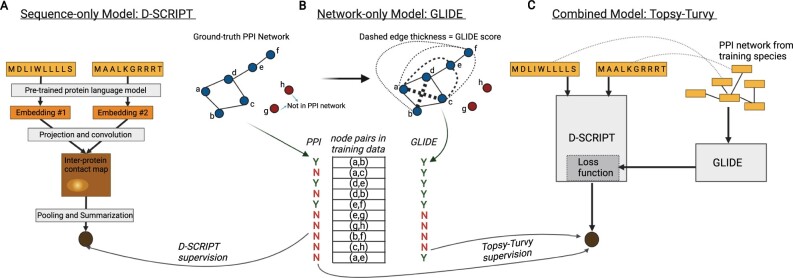
Fig. 1.
Topsy-Turvy synthesizes sequence-to-structure-based prediction using D-SCRIPT with network-based prediction using GLIDE. (A) D-SCRIPT uses a protein language model to generate representative embeddings of protein sequences, which are combined with a convolutional neural network to predict protein interaction. It is supervised using binary interaction labels from the training network and regularized by a measure of contact map sparsity. (B) GLIDE scores all possible edges using a weighted combination of global and local network scores which are learned from the edges already in the training network. (C) Topsy-Turvy is supervised with both the binary interaction labels of the true (training) network and with the GLIDE predicted scores, thus integrating bottom-up and top-down approaches for PPI prediction into the learned Topsy-Turvy model