Fig. 3.
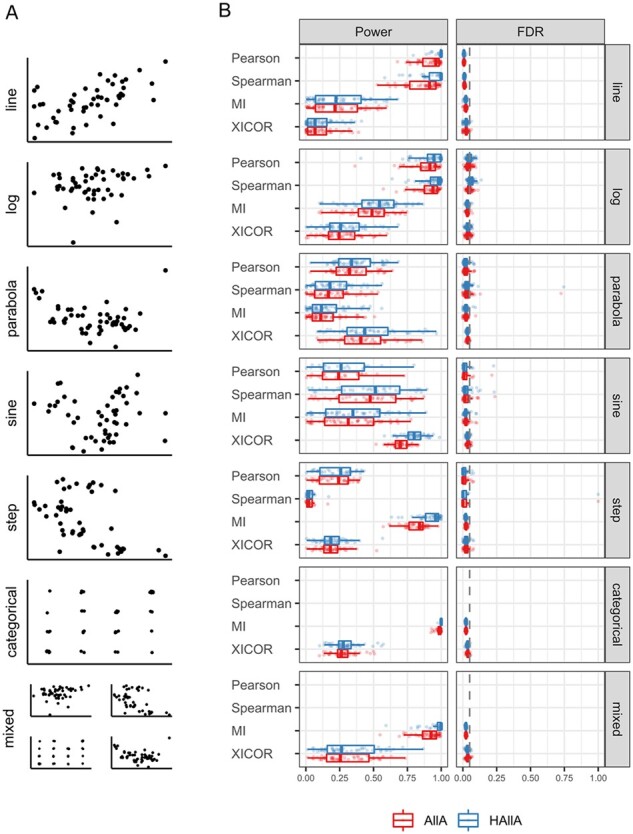
HAllA discovers block-structured associations while controlling FDR. (A) For a variety of feature linkage relationships, we simulated 50 independent paired datasets, each containing 200 features, 50 samples and clusters of correlated features. We then evaluated the ability of hierarchical versus AllA testing to recover these associations using a variety of similarity metrics. (B) Performance was evaluated by comparing power and FDRs. Our hierarchical AllA approach improved sensitivity relative to naive AllA approaches at a comparable FDR. Similarity metrics that do not accept categorical data have not been evaluated in the categorical or mixed association type. Other similarity metrics included in HAllA (dCor, NMI) were not applied in these simulations because their reliance on permutation tests made them too slow for simulations of this size (i.e. with many repeated iterations), although they are typically practical in individual real-world datasets
