Figure 3.
Human CCAN binds DNA
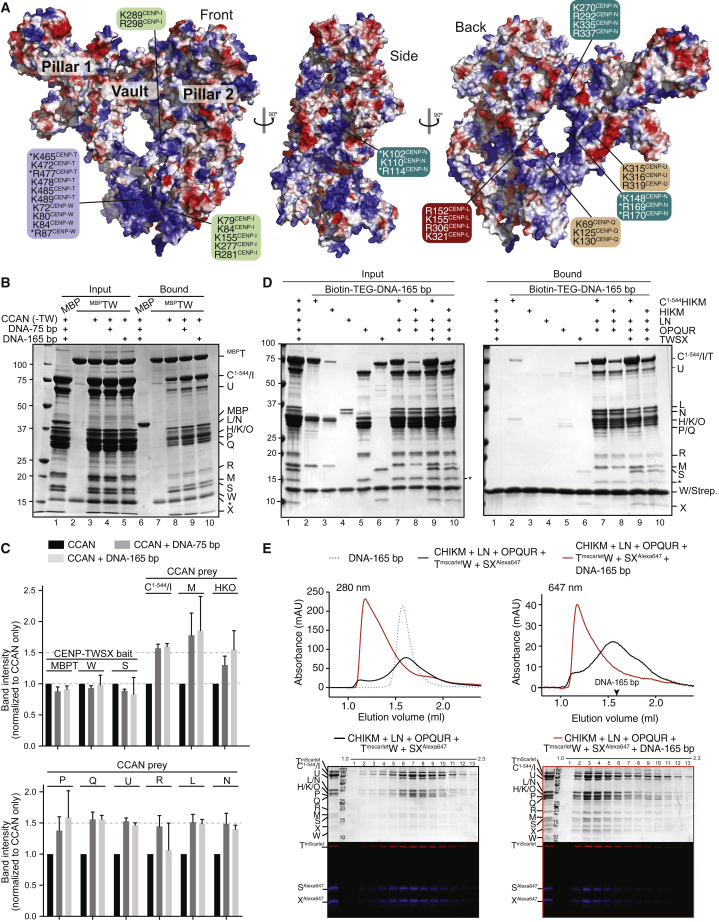
(A) Surface electrostatics (red, negative; blue, positive; potential display levels were between −80 and 80 kT/e) on human CCAN complex. Positively charged residues contributing to potential DNA-binding interfaces are indicated. Asterisks mark residues previously shown to affect DNA binding (Chittori et al., 2018; Nishino et al., 2012; Takeuchi et al., 2014; Zhou et al., 2021).
(B) Left: Binding assay on amylose beads with MBP (negative control) and MBPCENP-TW as baits. CCAN subunits were added in solution, with or without DNA. Beads were recovered by centrifugation, washed, and analyzed by SDS-PAGE. The asterisk here and in other panels with SDS-PAGE gels marks a proteolytic product of CENP-R (probably its folded core).
(C) Quantified band intensities of indicated subunits or group of subunits (when co-migrating) normalized to their intensity in the absence of DNA. The quantification reflects three technical replicates. Error bars indicate SD.
(D) Biotin-TEG (triethyleneglycol) DNA (165 bp) immobilized on streptavidin beads was incubated with the indicated CCAN subcomplexes. Inputs and bound proteins were visualized by SDS-PAGE and Coomassie staining.
(E) Size-exclusion chromatography of the indicated complexes with or without 165-bp dsDNA. Proteins separated by SDS-PAGE were visualized with Coomassie (top) or fluorescence (bottom). Profiles report absorbance at the indicated wavelengths.