Figure 5.
A yeast Ctf19CCAN:CENP-A nucleosome structure is a poor model for the human CCAN
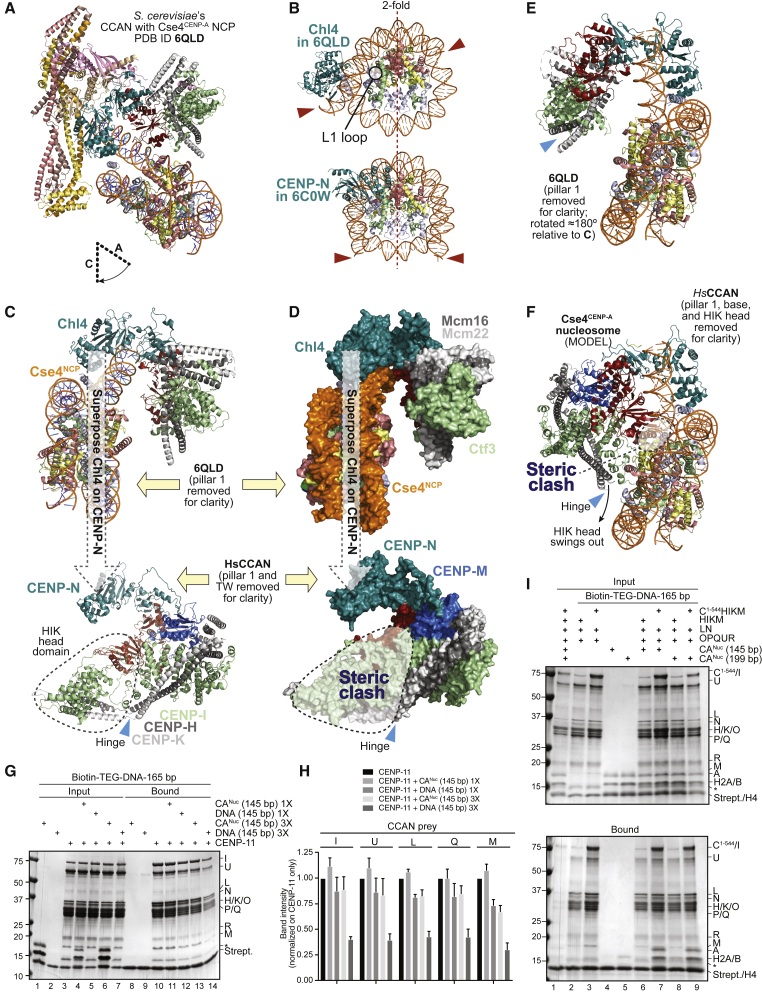
(A) Cartoon of the S. cerevisiae Ctf19CCAN:CENP-A nucleosome complex (PDB: 6QLD) (Yan et al., 2019).
(B) CENP-N is differently positioned on yeast and human nucleosome structures. The nucleosome cores are shown with the same orientation, indicated by the 2-fold pseudosymmetry axis. Red arrows point to DNA ends in the two structures. In 6QLD, the nucleosomal DNA unwraps from one end and is attracted into the vault. The ends are instead aligned with the 2-fold pseudosymmetry axis in 6C0W. H2A C-terminal tails were removed for clarity.
(C) Top: Rotated view of the yeast complex (6QLD) already shown in (A). Pillar 1 was removed for clarity. Chl4CENP-N was used to superpose Ctf19CCAN on human CCAN (bottom, pillar 1 and the base were also removed for clarity, only vault and pillar 2 are shown). The HIK head domain is boxed (dashed line).
(D) Objects in (C) are shown as surfaces. Superposition of CENP-NChl4 predicts a dramatic steric clash of the nucleosome with the HIK head domain.
(E) 6QLD is displayed as in (C) but rotated approximately 180°. There are no contacts of pillar 2 with the Cse4 nucleosome core.
(F) Human CCAN (same orientation to the yeast complex in E) was modeled onto 6QLD by aligning CENP-N on Chl4CENP-N. The predicted steric clash of HIK head with the Cse4CENP-A nucleosome (demonstrated in D) can be solved by a swinging-out rotation about the hinge. There are residual predicted clashes of CENP-L with H2A:H2B. Effects of the different relative position of pillar 2 relative to CENP-NChl4 in yeast and human are evident.
(G) CENP-11 was first immobilized by allowing its binding to biotinylated DNA on streptavidin beads. Free DNA or CENP-A nucleosomes at 1× or 3× ratio to protein were then added as indicated. Beads were washed and analyzed by SDS-PAGE.
(H) Three technical replicates of the experiment in (G) were quantified.
(I) The indicated CCAN species were immobilized on streptavidin beads coated with biotinylated DNA. Presence of CENP-C1–544 distinguished the CENP-11 and CENP-12 complexes. Retention of 145- or 199-bp CENP-A nucleosomes was only observed in presence of CENP-C. Three technical repeats were performed.