Figure 7.
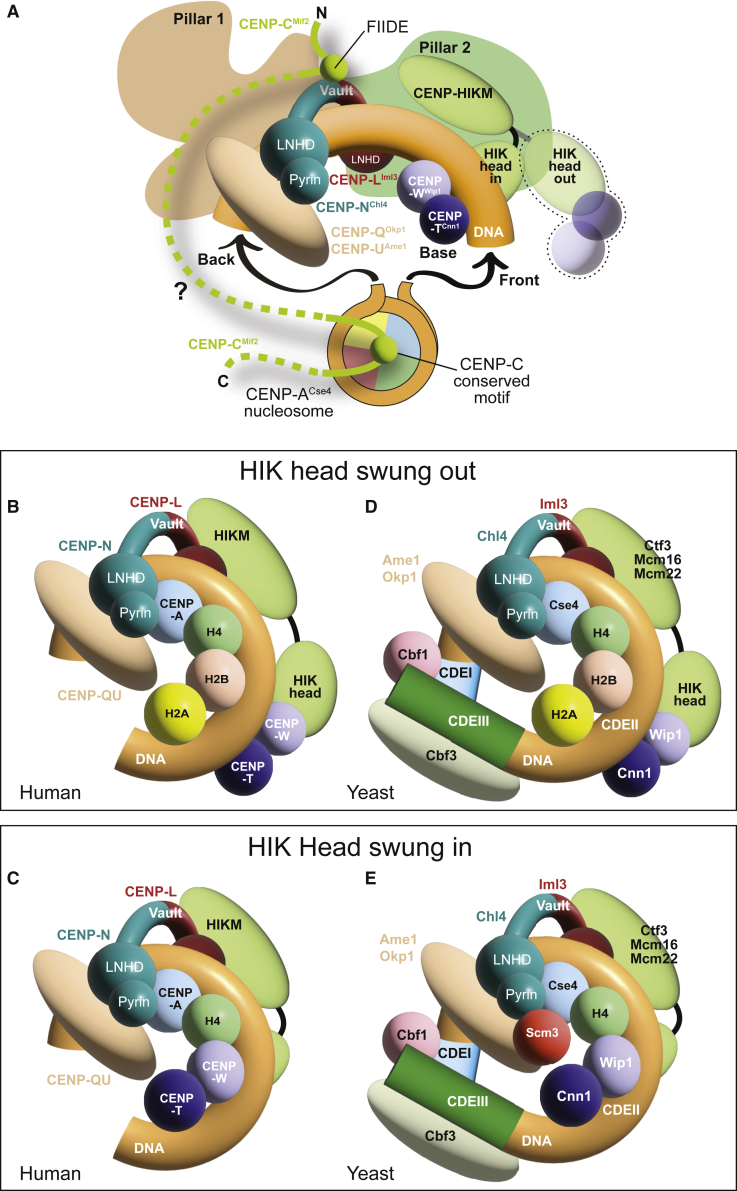
Models of centromere:chromatin interaction
(A) Common features of the CCANCtf19:CENP-ACse4 interaction in yeast and humans. The CENP-LIml3NChl4 vault is occupied by dsDNA that emerges from a CENP-ACse4 nucleosome that is otherwise not directly integrated in CCANCtf19 and only connected to it through CENP-CMif2, which acts as the crucial link between the nucleosome and the CCANCtf19. CENP-CMIf2 is flexible (dotted line), enabling multiple binding modes observed or predicted in yeast and humans. The conserved motif (and presumably the central region if present) binds the CENP-ACse4 nucleosome. The FIIDE motif is only detected in human CENP-C, but an equivalent region of Mif2CENP-C binds to Iml3CENP-L:Chl4CENP-N in S. cerevisiae (Hinshaw and Harrison, 2013). In the “swung-in” conformation, the HIK head positions the attached CENP-TCnn1WWip1 in the observed “base” position (this conformation would not be available to the yeast complex due to the divergence of pillar 2). A putative “swung-out” conformation is also shown.
(B) In this model CENP-A:H4 faces the pyrin domain of CENP-N. A single filament of dsDNA is allowed inside the CENP-LN vault. The CENP-TW base connected to the HIK head is in a swung-out conformation that permits an interaction of CENP-A:H4 with H2A:H2B, with which CENP-TW would otherwise clash. CENP-QU may contribute, alone or in complex with other proteins, to prevent CENP-A dimerization, generating a hemisome.
(C) The same hemisome complex, but with H2A:H2B replaced by CENP-TW as expected for the swung-in conformation observed in our structures.
(D) The CDEII core of the yeast point centromere is ∼85 bp long, and Cse4CENP-A is precisely positioned on it (Cole et al., 2011; Furuyama and Biggins, 2007). The CDEII core is flanked by CDEI and CDEIII motifs that associate with Cbf1 and Cbf3. A hemisome model has been proposed for this organism (see main text). As shown, the wrap of the DNA in the model is left-handed, but there is evidence for right-handedness (Diaz-Ingelmo et al., 2015; Furuyama and Henikoff, 2009; Huang et al., 2011).
(E) Another hemisome model may explain depletion of H2A:H2B at yeast centromeres as well as a function of Scm3 in preventing Cse4CENP-A dimerization. This swung-in conformation of the HIK arm would require a large-scale conformational change of pillar 2, making it resemble the human complex.