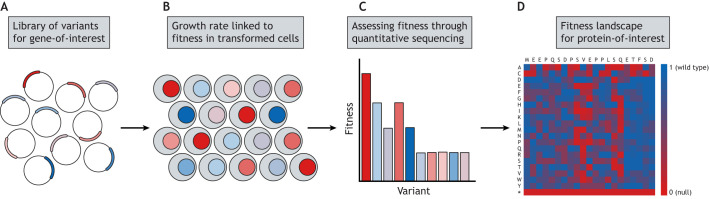
Fig. 2.
Summary of a typical DMS experiment. (A) A library of variants, often representing every possible amino acid substitution in a protein, is generated and cloned into expression vectors. (B) The vectors are then introduced to mammalian or yeast cells where the function of the mutant protein is linked to the cell growth rate or some other measurable attribute. (C) Variant fitness is measured at different time points by quantitative sequencing, and compared to positive and negative controls to calculate relative fitness values. (D) A fitness map of all possible variants in the protein can be constructed from the relative fitness data.