Figure 2.
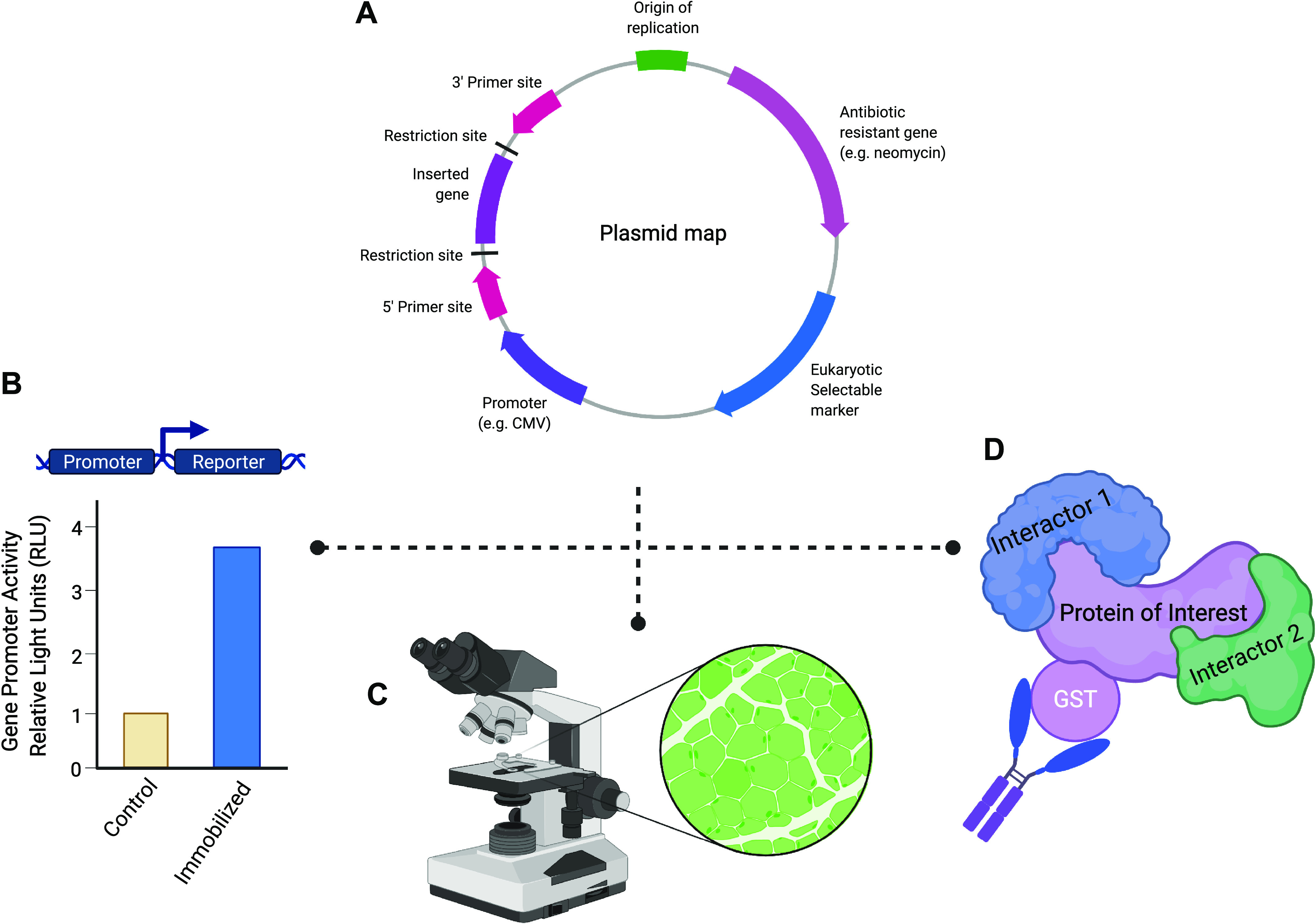
Applications of plasmid DNA into skeletal muscle physiology research. A: use of mammalian expression plasmids can aid in mechanistic experiments because they can be introduced by transfection (e.g., electroporation) into host cells. Expression plasmids possess a multiple cloning site (MCS) immediately downstream of a promoter sequence that allows for the amplified cDNA of a gene of interest to be cloned into the plasmid. The MCS of a plasmid consists of numerous unique restriction enzyme recognition sequences that simplify the cloning process and allow for the proper directional insertion of the cDNA of a gene of interest. Furthermore, the use of a viral promotor (e.g., CMV or SV40) allows for robust expression of the target gene within eukaryotic cells. Finally, the cDNA cloned into an expression plasmid can be easily sequenced using commonly used primer sites (e.g., T7, SP6, and T3) to confirm successful target gene insertion and to verify that no mutations have been introduced during the cloning process. B: transcriptional regulation of a gene in skeletal muscle under various stimuli can be ascertained by using a reporter plasmid construct containing the promoter of a gene of interest inserted upstream of a reporter gene (e.g., luciferase). For example, in immobilized/disuse muscle atrophy, the target gene promoter activity can be measured in control and immobilized tissues that have been transfected with the reporter plasmid construct. Under an atrophy stimulus, the gene of interest promoter activity will be detected through changes in the amount of luciferase activity. C: use of a fluorescent reporter, such as green fluorescent protein (GFP), allows for the identification of successfully transfected muscle fibers under the microscope. The incorporation of a GFP-tag into a target protein can aid in studying the localization of novel proteins in skeletal muscle fibers. D: to investigate protein-protein interactions and complexes, epitope tags [e.g., hemagglutinin (HA), FLAG, myc, and glutathione S-transferase (GST)] provide a useful tool that can be used in common molecular biology techniques such as coimmunoprecipitation to identify novel interactors of the protein of interest. Most commonly, the epitope tag is integrated into the plasmid sequence so that the tag will be easily added in frame with either the C- or N-terminus of the target protein. In addition, these tags can be used to identify proteins when a specific antibody against a protein of interest is not yet available. Created with Biorender.com.
