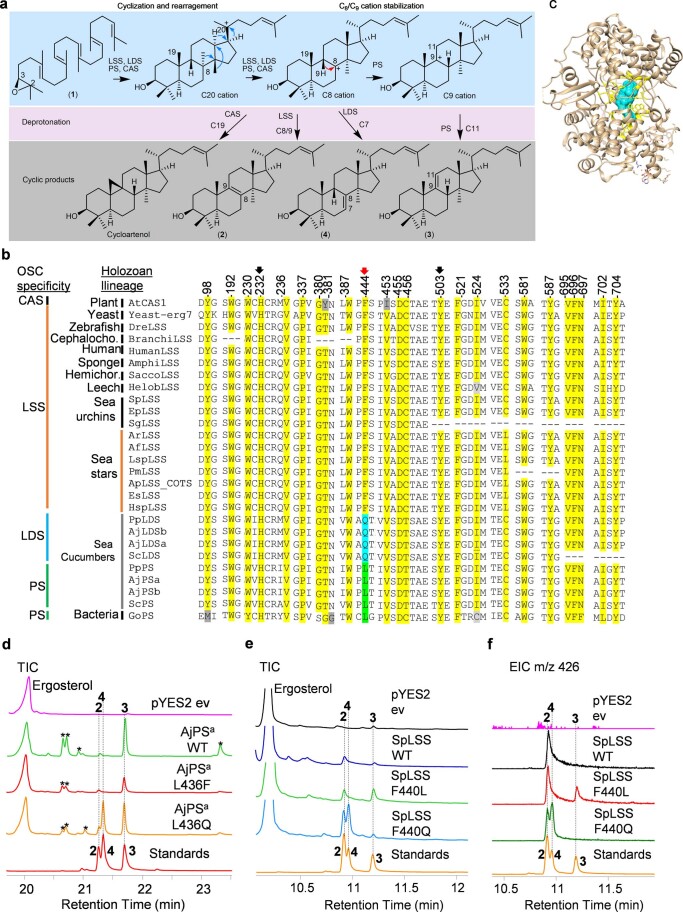
Extended Data Fig. 8. A single active site residue (position 444) determines cyclization mechanism and product specificity of LSS, LDS and PS OSCs.
a, The cyclization mechanism for lanosterol (2), lanostadienol (4), parkeol (3) and cycloartenol. Protonation, cyclization, and rearrangement of 2, 3 oxidosqualene (1) to a central protosteryl cation (C20). Rearrangement of C20 cation lead to either C8 or C9 cations depending on type of OSC involved. Deprotonation at C7 of C8 cation lead to lanostadienol (4) and deprotonation at C11 of C9 cation lead to parkeol (3). Lanosterol (2) and cycloartenol are derived from C8 and C9 cations, respectively. b, OSC sequence alignment of 24 active site residue positions of echinoderms and others (sequences given in Supplementary Notes 2). c, OSC active site region is shown in yellow and region encompassing 5Å around lanosterol is in blue. d, GC-MS TICs of yeast extracts expressing AjPSa-WT and its mutants AjPSa-L436F, AjPSa-L436Q. Peaks marked with asterisks are undesired modifications of the PS product in yeast. e-f, TICs and EICs (m/z 426) of yeast extracts expressing SpLSS-WT and its mutants SpLSS-F440L, SpLSS-F440Q. Standards: 2, lanosterol; 4, lanostadienol; 3, parkeol.