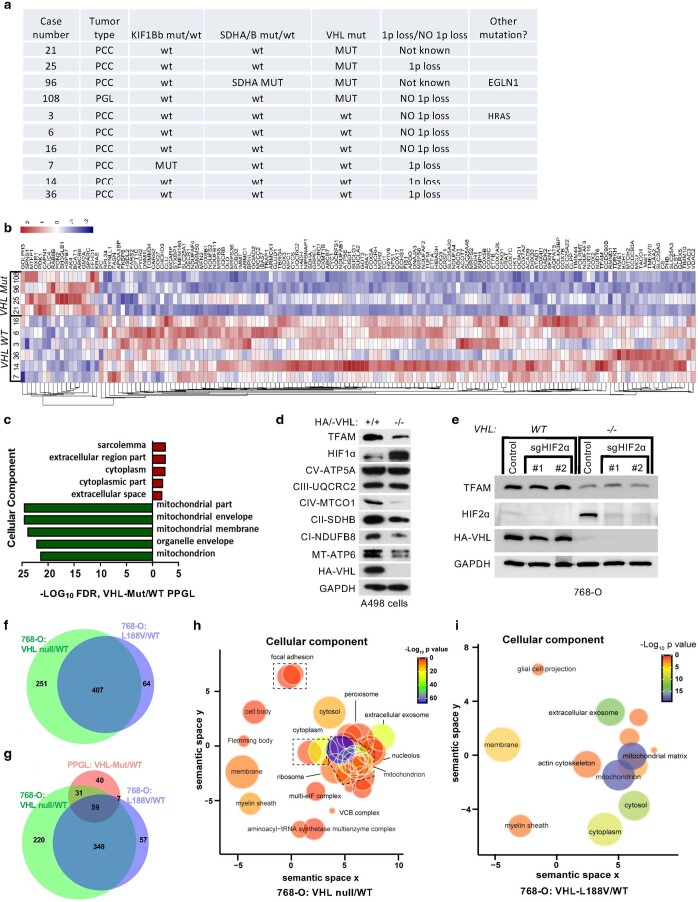
Extended Data Fig. 1. VHL regulates of mitochondrial mass independent of HIFa.
(a) List human primary PPGL tumors with characterized mutation status and 1p36 status that were analyzed by by nanoLC-MS/MS in Fig. 1A-D. wt = wild-type. (b) Heatmap of significantly regulated mitochondrial proteins in VHL-mutant compared to VHL wild-type PPGL tumors (p < 0.05, two-tailed unpaired t test). (c) Top 5 cellular component of top 50 up (red)- and down (green)-regulated proteins for human VHL mutant PCC/PGL tumors compared to VHL wild type PCC/PGL tumors according to the false discovery rate (FDR). Medium confidence threshold (0.4) was used to define protein-protein interactions. (d) Immunoblot analysis of A498 VHL-null cells (−/−) stably transfected to generate HA-VHL (WT). n = 3 biological independent experiments. (e) Immunoblot analysis of 786-O cells with indicated genotype stably transduced with lentivirus encoding sgRNA targeting HIF2α. n = 3 biological independent experiments. (f) Venn diagram representing significantly downregulated proteins shared in VHL-null 786-O cells with type 2 C VHL-L188V mutant cells and (g) shared with VHL mutant PPGL. (h, i) GO term enrichment in cellular component of 393 significantly down-regulated proteins (p values < 0.0001, two-tailed unpaired t test) comparing VHL-null to VHL-WT cells (h) and 200 significantly down-regulated proteins (p values < 0.0001, two-tailed unpaired t test) comparing VHL-L188V to VHL-WT cells (i) performed using DAVID and plotted using REVIGO. The size of the bubbles is indicative of the number of proteins annotated with that GO term; bubbles are color coded according to significance.