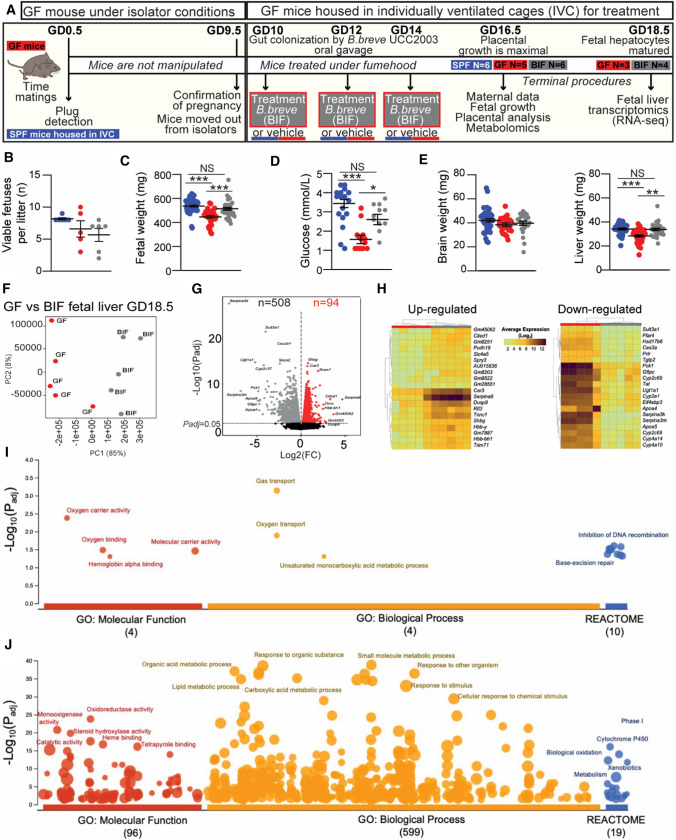
Fig. 1.
Effects of the maternal gut microbiome and B. breve supplementation during pregnancy on fetal viability, growth and hepatic transcriptome. A Experimental design. B Number of viable fetuses per litter (One-way ANOVA with Tukey’s multiple comparison). C Fetal weight. D Circulating glucose concentrations in fetal blood. E Fetal organ weights. F–G RNA-Seq analysis of fetal liver samples obtained at GD18.5. F PCA plot and G volcano plots showing up and down-regulated differentially expressed genes (DEGs) in BIF group (compared to GF group). H Heat map of the 20 most up and down-regulated DEGs (BIF group). I, J Functional profiling (g:Profiler) on 602 DEGs. Key enriched GO terms and REACTOME pathways are shown in the figure. Fetal data are obtained on GD16.5 from: SPF (49 fetuses/6 dams), GF (33 fetuses/5 dams), BIF (34 fetuses/6 dams). Dots represent raw data for each variable assessed (individual values). However, the statistical analysis and the mean ± SEM reported in the graphs were obtained with a general linear mixed model taking into account viable litter size as a covariate and taking each fetus as a repeated measure followed by Tukey multiple comparisons test (further explanations can be found in the Materials and Methods, statistical analysis section). Identification of outliers was performed with the ROUT Method. RNA-seq was performed on fetal livers obtained at GD18.5 from a total of 3 GF and 4 BIF pregnant dams/litters. RNA-Seq data analysis is described in the material and methods section (NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001)