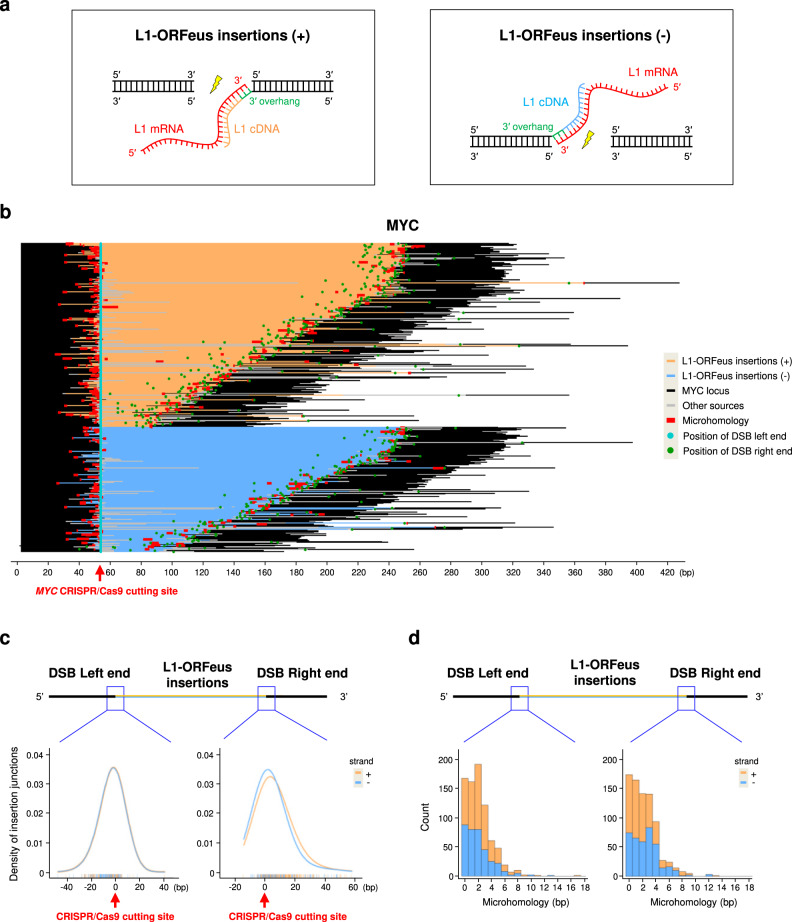
Fig. 2. Detailed characterization of de novo L1-ORFeus insertions at CRISPR/Cas9 target sites.
a A schematic model of L1-ORFeus insertions at either DNA end of the CRISPR/Cas9-mediated DSB. CRISPR/Cas9 cutting and subsequent end-resection provide a 3′overhang (green) that can be primed by 3′ end of L1-ORFeus mRNA (red) for retrotranscription to synthesize the L1 cDNA. The L1 insertions (+) in orange represents the orientation of sequenced L1-ORFeus sequence is from 5′ to 3′ (a, left), while (−) in blue represents the orientation of sequenced L1-ORFeus sequence is from 3′ to 5′ (a, right). b L1-ORFeus junction analysis obtained by amplicon sequencing at the MYC locus targeted by MYC CRISPR/Cas9 in HEK293T cells expressing L1-ORFeus. The sequence alignment was centered on the left end of CRISPR-Cas9-mediated DSBs. Other sources include genomic fragments or plasmid insertions. c Density and distribution of L1-ORFeus insertion junctions pooled from MYC (b) and RAG1 (Supplementary Fig. 2i). The bar in the bottom represents the position of the insertion junction that join to the left or right end of the CRISPR/Cas9-mediated DSB in HEK293T cells expressing L1-ORFeus. The orientation of L1-ORFeus insertion junction is shown in orange when the fragment is oriented 5′ to 3′ (+) or blue when 3′ to 5′ (−). d Histogram plots of microhomology lengths in junctions (from c) joining L1-ORFeus to the left or right end of the CRISPR/Cas9-mediated DSB in HEK293T cells expressing L1-ORFeus. The orientation of L1-ORFeus insertions is shown in orange when the fragment is oriented 5′ to 3′ (+) or blue when 3′ to 5′ (−).