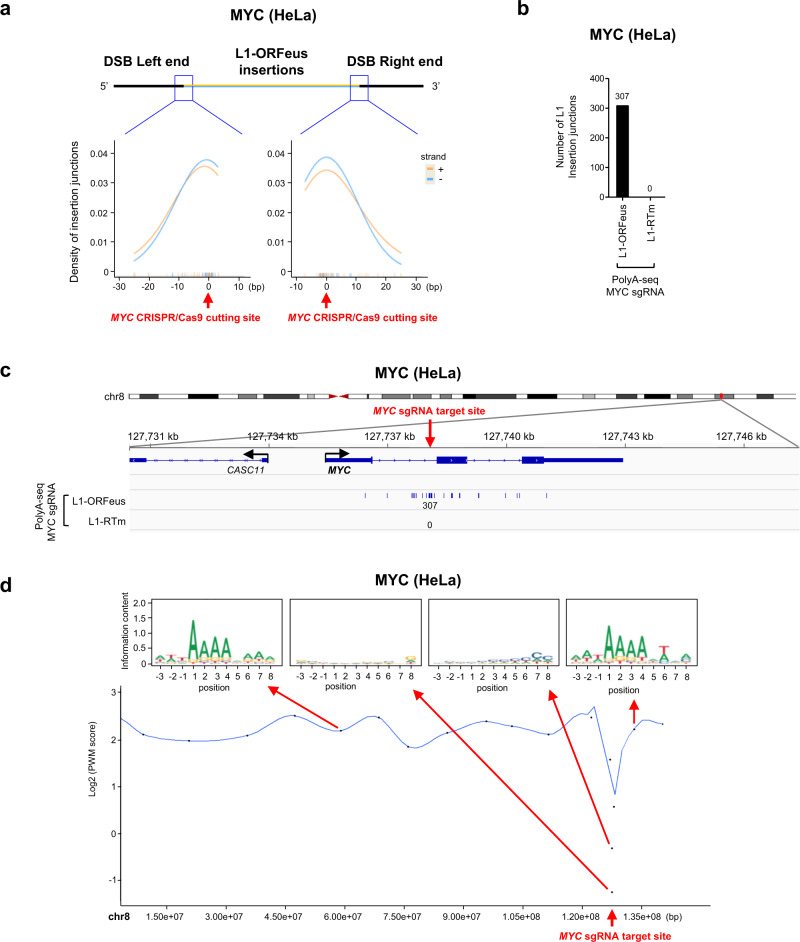
Fig. 5. RT-dependent de novo L1-ORFeus insertions at CRISPR/Cas9 target site in HeLa cells.
a Detailed view of the distribution of L1-ORFeus insertions at the left or right end of the MYC locus obtained by amplicon sequencing at the MYC locus targeted by MYC CRISPR/Cas9 in HeLa cells expressing L1-ORFeus. b Numbers of L1-ORFeus insertions at the MYC locus obtained by PolyA-seq targeted by MYC CRISPR/Cas9 in HeLa cells that express L1-ORFeus or L1-RTm. c Detailed view of the distribution of L1-ORFeus insertions at the MYC locus obtained by PolyA-seq in HeLa cells targeted by MYC CRISPR/Cas9 expressing L1-ORFeus or L1-RTm. d Top: Sequence logo representing the consensus motif detected at L1-ORFeus pre-integration sites in example regions proximal or distant to the MYC CRISPR/Cas9 DSB. Each dot represents a chromosomal region encompassing 30 independent L1 insertions. Bottom: Spline interpolation curve of consensus motif position-weighted matrix (PWM) score in chromosome 8 obtained by PolyA-seq in HeLa cells targeted by MYC CRISPR/Cas9 that express L1-ORFeus. The mean interval size covered by the chromosomal region encompassing 30 independent L1-ORFeus insertions was: 235 bp (range 28–442 bp) within approx. ±5 kb of the MYC CRISPR/Cas9 target site, and 8,025,664 bp (range 863,222–16,390,320 bp) outside of the CRISPR/Cas9 target site.