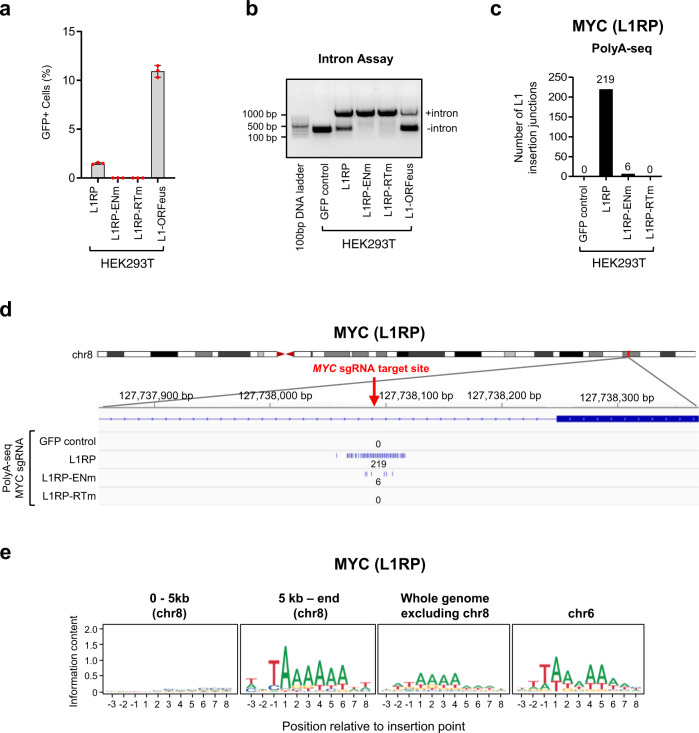
Fig. 6. RT-dependent de novo L1RP insertions at CRISPR/Cas9 target site in HEK293T cells.
a The percentage of GFP-positive HEK293T cells was analyzed by FACS at day 11 after L1RP transfection and CRISPR/Cas9-mediated DSBs at the MYC locus as depicted in Supplementary Fig. 8b. Data are mean ± SD, n = 3 independent experiments. b PCR assay across the intron of the GFP reporter gene (Intron assay) was performed to probe the L1RP retrotransposition events by genomic DNA-PCR. HEK293T cells transfected with the same amount (1 μg) of L1-ORFeus reporter served as a control. c Numbers of L1RP insertions obtained by PolyA-seq at the MYC locus targeted by MYC CRISPR/Cas9 in HEK293T cells expressing L1RP, L1RP-ENm, L1RP-RTm, or GFP control. d Detailed view of the distribution of L1RP insertions obtained by PolyA-seq at the MYC locus targeted by MYC CRISPR/Cas9 in HEK293T cells expressing L1RP, L1RP-ENm, L1RP-RTm, or GFP control. Each line corresponds to an independent L1RP insertion; number of L1RP insertions is pooled from four independent PolyA-seq experiments. Red arrow shows the MYC CRISPR/Cas9 target site. e Sequence logos representing the consensus motif detected at L1RP pre-integration sites in the indicated regions proximal or distant to the MYC CRISPR/Cas9 DSB in chromosome 8. The canonical consensus motif in chromosome 6 served as a control. Source data are provided as a Source data file.