FIGURE 1.
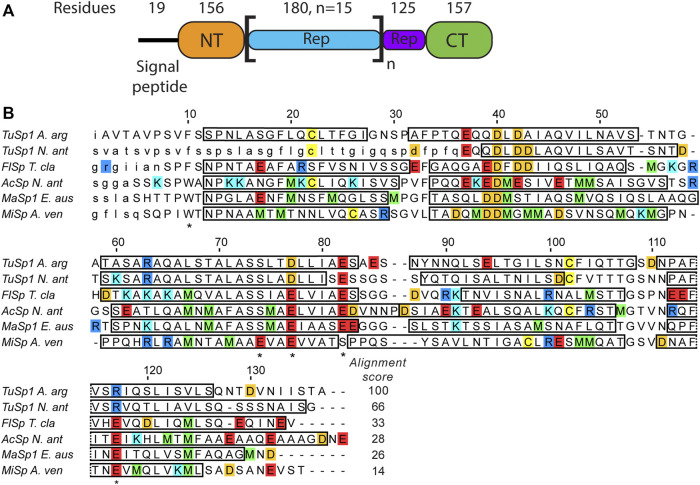
(A) Argiope argentata TuSp domain architecture (Chaw et al., 2018). The number of residues for NT is different from the construct used in this study because it includes ∼20 non-conserved residues linking it to the Rep region. (B) Amino acid sequence alignment of spidroin NTs with known tertiary structures. Included are NTs from Argiope argentata TuSp (PDB ID 6TV5), Nephila antipodiana TuSp (PDB ID 2K3Q), Nephila clavipes FlSp (PDB ID 7A0O), Nephila antipodiana AcSp (PDB ID 7BUT), Euprosthenops australis MaSp (PDB ID 2LTH) and Araneus ventricosus MiSp (PDB ID 2MX9). Locations of α-helices are indicated by rectangles and amino acids excluded from the expression vector are in small caps. Residues protonated upon dimer formation (E79, E84, E119 in MaSp and in MiSp E73, E79 and E119) and W10/F10 are marked with an asterisk below the sequence. Arginines are blue, lysines are cyan, glutamic acids are red, aspartic acids are orange, methionines are green and cysteines are yellow. ClustalW alignment score to E. australis MaSp NT is shown at the end of each sequence.