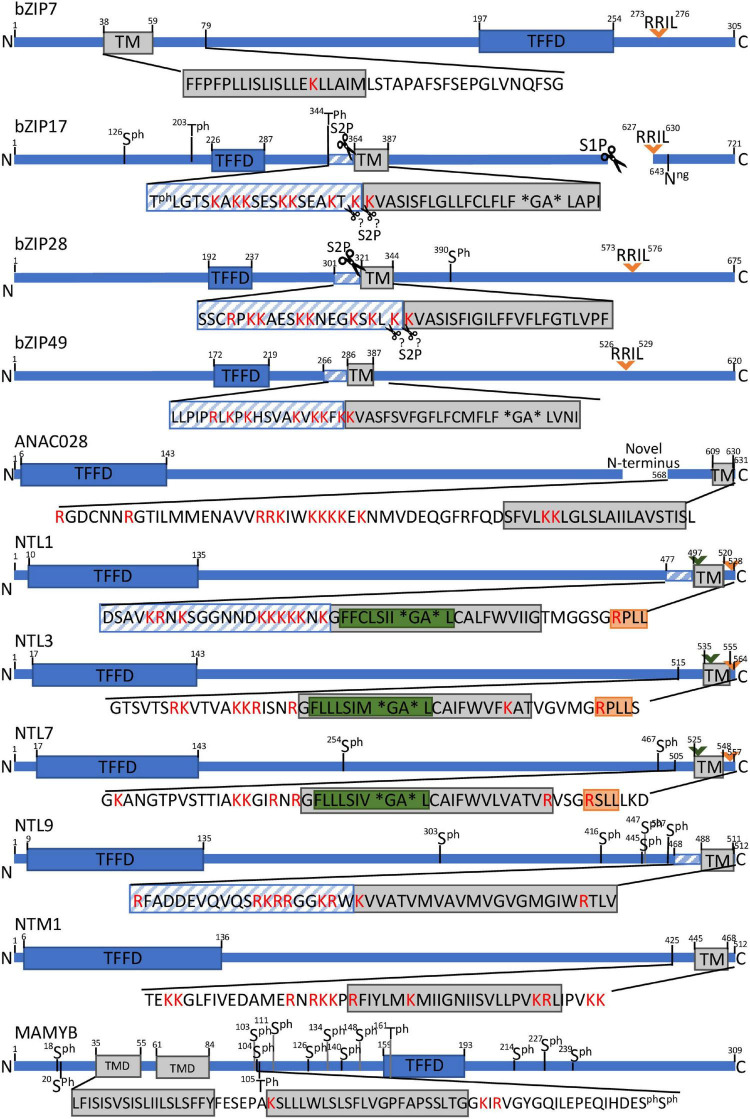
FIGURE 2.
Schematic presentation of protease recognition motifs and structural features indicative of RIP activation for a selection of MB-TFs. Predicted rhomboid recognition sites (LxLSIxGA) are indicated in green and (predicted) SIP recognition sites (Rx[LIT][KL]) in orange. Diagonal blue stripes mark TMD-neighboring regions that are significantly (corrected P value < 0.05) enriched for positive amino acids (R and K. lysine and arginine) and scissors indicate known or predicted cleavage sites. Asterisks specify helix-breaking residues. Ph. phosphorylated amino acid.