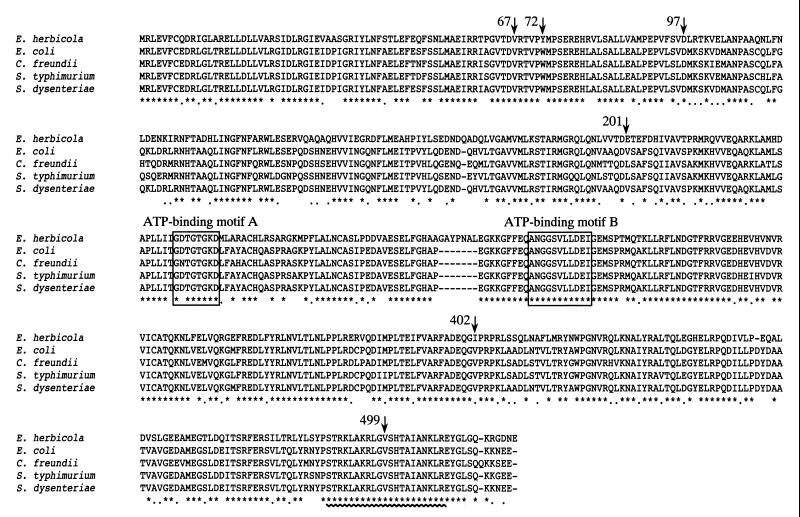
FIG. 1.
Multiple amino acid sequence alignment of TyrR proteins. The deduced amino acid sequence of the E. herbicola TyrR protein was aligned with those of the TyrR proteins from E. coli (GenBank accession number M12114) (9), S. enterica serovar Typhimurium (S. typhimurium) (GenBank accession number U90141) and C. freundii (GenBank accession number U90140) (Bai and Somerville, unpublished data), and S. dysenteriae (GenBank accession number AF153317) (38) by use of the Clustal W 1.6 program (51). The asterisks indicate residues that are conserved in all five TyrR proteins, and the dots indicate positions at which only conservative changes have occurred. Only the E. herbicola TyrR protein possessed seven extra amino acid residues between the two ATP-binding motifs (A and B), which are enclosed by boxes. The sequences in the HTH motifs are identical in all five proteins and are underlined with a wavy line. Mutations mapped in this experiment are indicated by arrows labeled with their amino acid positions.