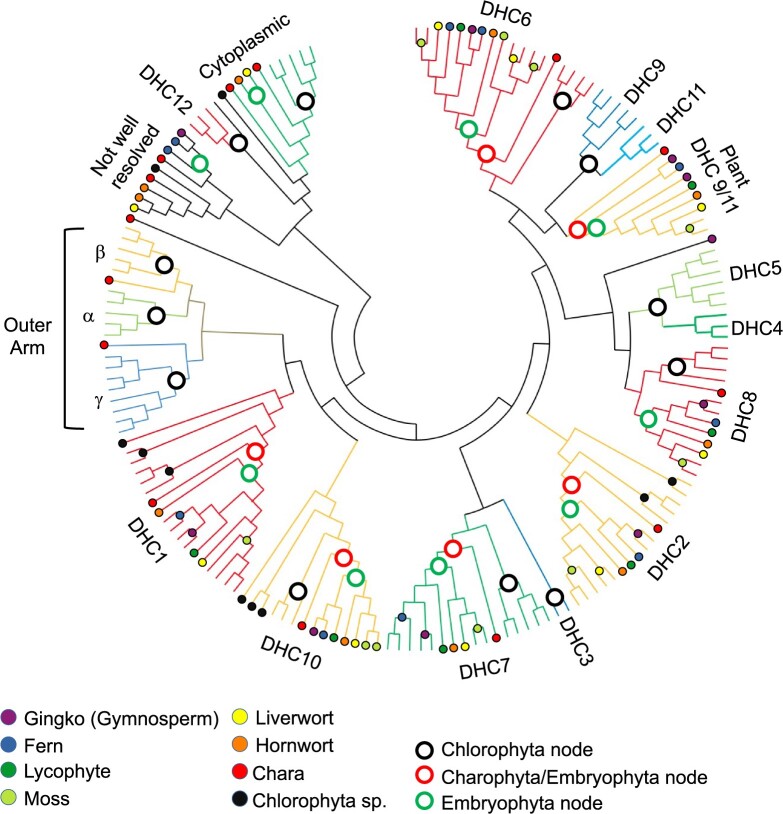
Figure 2.
Topology of maximum parsimony tree showing DHC subcategories in Viridiplantae and humans. Amino acid sequences of 169 DHCs were sorted into classes by maximum-parsimony analysis of their evolutionary history. Groups in bold text are found in all plants with flagellated sperm and asterisks on DHC12 and DHC16 indicate these are found only in hornworts and liverwort but not other land plants. Three outer arm dynein sequences are found in C. reinhardtii: α, β, and γ. Additional moss (Ceratodon purpureus and Sphagnum magellanicum) genomes were searched to corroborate this loss. Proteins were classified based upon the identity of Homo sapiens and the C. reinhardtii protein within each color-coded clade (human sequences are noted with a black dot for comparison). Sequences were aligned in ALIVIEW and analyzed using MEGAX (Kumar et al., 2018). Sequence fragments less than 1.5 kb were removed prior to alignment. All sites with less than 50% coverage were eliminated leaving a total of 3,766 positions. The bootstrap consensus tree topology of 500 replicates is presented using circular format. A linear format version of this tree including individual sequence accessions and bootstrap replicate values is included as Supplemental Figure S1.