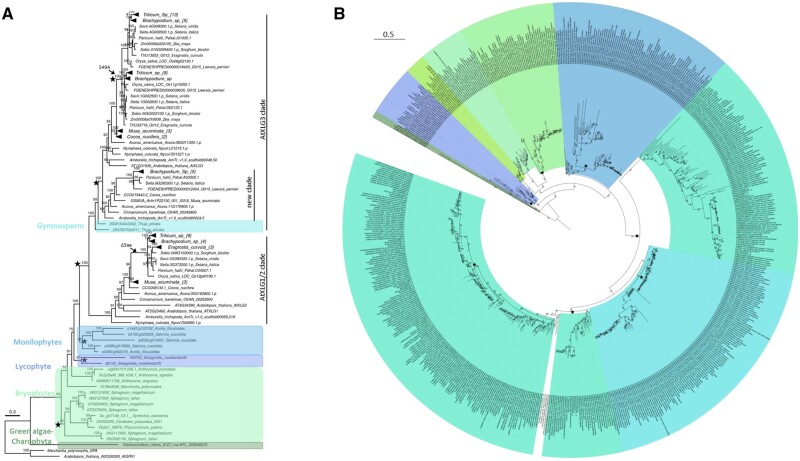
Figure 3.
Phylogenetic analysis of plant and algal XLGα proteins. Maximum likelihood tree of XLGα proteins from all plant and algal species, excluding eudicots. A, Tree of XLGα sequences from publicly available genomes. Plant Gα sequences are used as outgroup. Many nodes are collapsed and labeled with triangles to improve readability. The total number of sequences in each collapsed node is mentioned in their respective tip label inside the curly bracket. Tree scale 0.3 amino acid substitution per site. B, Tree of XLGα sequences from publicly available genomes and transcriptomes. Tree scale 0.5 amino acid substitution per site. In both trees, each plant lineage is highlighted and labeled in different colors. Bootstrap support values above 40% are shown at each node (1,000 bootstrap replicates). Star marks the gene duplication events inferred from the tree topology.