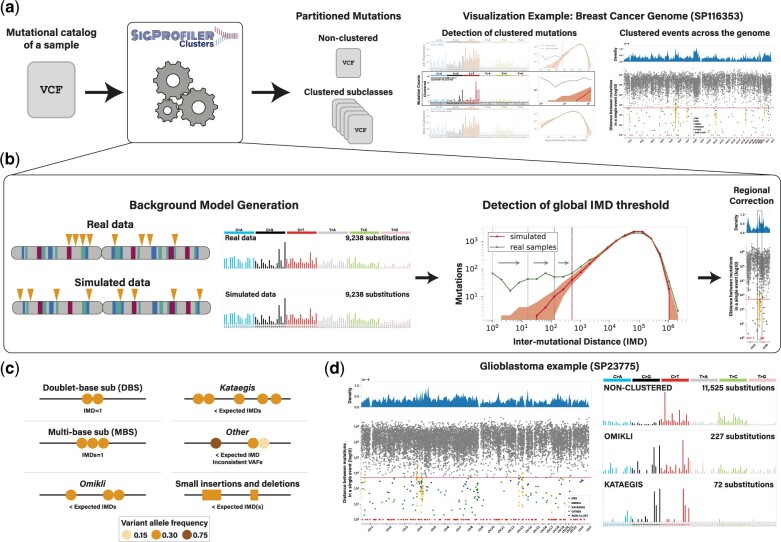
Fig. 1.
Detection and characterization of clustered mutations with SigProfilerClusters. (a) An example workflow used to detect clustered mutations in a single cancer genome. As an input, SigProfilerClusters accepts common formats for mutations, such as ones in the variant calling format (VCF), and the tool separates all clustered mutations from the complete mutational catalog of the provided sample. Final partitions of mutations in the sample are outputted as VCF files and visualized using the mutational spectra of all mutations, only clustered mutations and only non-clustered mutations along with a rainfall plot commonly used to show the distribution of inter-mutational distances across a cancer genome (Alexandrov et al., 2013; Bergstrom et al., 2022; Nik-Zainal et al., 2012). (b) Schematic demonstrating the process of calculating a sample-dependent IMD threshold to separate clustered from non-clustered mutations across each genome. A binary search algorithm is used to efficiently detect the optimal global IMD threshold for each sample. Detection of the global IMD threshold is illustrated using gray arrows. Regional corrections are performed to identify local IMD thresholds based on variance of mutation rates across the genome. (c) Every clustered mutation is classified into a single subcategory of clustered event. (d) Rainfall plot illustrating the distribution of IMDs across a single glioblastoma sample (left). The mutational spectra for omikli and kataegic events reveal a different mutational pattern compared to the pattern of all non-clustered somatic mutations (right)