Abstract
T cells engineered to express artificial chimeric antigen receptors (CARs) that selectively target tumor-specific antigens or deleterious cell types offer transformative therapeutic possibilities. CARs contain an N-terminal extracellular antigen recognition domain, C-terminal intracellular signal transduction domains, and connecting hinge and transmembrane regions, each of which have been varied to optimize targeting and minimize toxicity. We find that a CD22-targeting CAR harboring a CD8α hinge (H) exhibits greater cytotoxicity against a low antigen density CD22+ leukemia as compared to an equivalent CAR with a CD28 H. We therefore studied the biophysical and dynamic properties of the CD8α H by nuclear magnetic resonance (NMR) spectroscopy. We find that a large region of the CD8α H undergoes dynamic chemical exchange between distinct and observable states. This exchanging region contains proline residues dispersed throughout the sequence that undergo cis-trans isomerization. Up to four signals of differing intensity are observed, with the most abundantly populated being intrinsically disordered and with all prolines in the trans isomerization state. The lesser populated states all contain cis prolines and evidence of local structural motifs. Altogether, our data suggest that the CD8α H lacks long-range structural order but has local structural motifs that transiently exchange with a dominant disordered state. We propose that structural plasticity and local structural motifs promoted by cis proline states within the CD8α H are important for relaying and amplifying antigen-binding effects to the transmembrane and signal transduction domains.
Keywords: CD8, CAR T-cell, linker, intrinsically disordered proteins, proline isomerization
1. Introduction
Adoptive T-cell transfer offers promise of a cancer treatment protocol without damage to healthy tissues [1]. Tumor-specific targeting with long-term anti-cancer outcome has been achieved by engineering patient T cells to express artificial chimeric antigen receptor (CAR) transmembrane proteins at the cell surface. CAR expression renders the patient’s T-cells specific to tumor-specific antigen, resulting in the adoptive transfer of engineered lymphocytes with cytotoxic potential against malignant and/or other target cells [2]. This approach has been successful in the treatment of several cancers, including relapsed or refractory non-Hodgkin lymphoma [3, 4], multiple myeloma [5], and relapsed or refractory B-cell acute lymphoblastic leukemia [6–15]. Indeed, anti-tumor activity has been achieved against several hematological antigens, including CD19 [16], CD22 [11], CD20 [17], CD30 [4], B cell maturation antigen [8], as well as the simultaneous targeting of two of these antigens [10, 18–20]. Nevertheless, despite the ability to engineer CARs against solid tumor antigens, the success of CAR T-cell therapy in these tumors has been limited, in part due to challenges in CAR-T trafficking and function in the hostile intratumoral microenvironment [21, 22].
CARs contain extracellular antigen recognition and intracellular signal transduction domains that are connected by a hinge (H) and transmembrane (TM) domain. The antigen recognition domain is typically a single chain fragment variable (scFv) region derived from a monoclonal antibody, resulting in a specific interaction with the targeted epitope. Following the binding of a CAR T cell to a tumor cell, the intracellular CAR signaling domain(s) activates the T cell, inducing tumor cell destruction and CAR T cell proliferation. There have been many studies aimed at optimizing CAR binding and signal transduction by the intracellular domain [23–25], but the H and TM regions can also be optimized. Cell surface CAR expression and stability are affected by the TM and the H regulates CAR signaling threshold [26–30]. CD19-targeting CARs including either the CD8α or CD28 H and TM have been FDA approved. Indeed, CD8 and CD28 H-TM are present in CTL-019 (KYMRIAH, Novartis Pharmaceuticals Corp.) and axicabtagene ciloleucel (YESCARTA, Kite Pharma, Inc.), respectively. In the context of anti-CD19 CARs, the CD8α H was associated with lower toxicities compared to the CD28 H [31]. Importantly though, the dynamic nature of the H region has stymied its characterization although several CAR antigen recognition domain structures and a limited number of transmembrane domain structures are available [32–35].
Here, we compared the efficacy of a CD22-targeting CAR incorporating CD28 as compared to CD8α H and TM. The CD8α H resulted in improved cytotoxicity against leukemic cells with low CD22 antigen density. We then applied various biophysical techniques to characterize the structural and dynamic properties of the CD8α H. This H has dispersed prolines that we find to undergo cis-trans isomerization and promote dynamic exchange throughout much of the CD8α H.
2. Materials and Methods
2.1. CAR construct generation
Golden gate assembly was utilized to create each of the indicated CAR constructs. All enzymes and reaction buffers were purchased from New England Biolabs (Ipswich, MA, USA) and all primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA). Briefly, an anti-CD22 CAR construct using the m971 scFv for CD22 targeting [36] in a lentiviral backbone [37] was used as a template for creating CAR parts. The backbone for the modular CARs was created via NheI and SalI double digestion, run on an agarose gel, and recovered using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA). Primers containing BsmBI restriction sites with programmable overhangs specifying orientation and position were used for PCR generation of CAR fragments. Residual template plasmid was removed via DpnI digestion and plasmid was isolated using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). One-pot reactions for each construct, consisting of DNA template, modular regions, BsmBI-v2, T4 ligase, and T4 ligase buffer, were performed in a Pro Flex PCR system (Thermo Fisher, Waltham, MA, USA) as described by the manufacturer [38]. The resulting reaction mixture (4µL) was used to transform One Shot™ Stbl3™ Chemically Competent E. coli (Thermo Fisher). Single colonies were picked from LB agar plates, cultured overnight, and plasmid DNA was purified using the ZR Plasmid Miniprep-Classic (Zymo Research). Constructs were sequence verified by Sanger sequencing Psomagen, Rockville, MD, USA) and DNA generated using the NucleoBond Xtra Midi kit for transfection-grade plasmid DNA (Macherey-Nagel, Düren, Germany).
2.2. Lentivirus production and generation of CART cells
The Lenti-X 293T lentiviral packaging cell line (Takara Bio, Kusatsu, Shiga, Japan) was co-transfected with each CAR construct, the packaging plasmid, Rev plasmid, and VSV-G pseudotyping envelope plasmid, as previously described [39]. Human T cells were transduced as previously described [39] and CAR surface expression was assessed using a CD22-Fc fusion protein (R&D Sytems, Minneapolis, MN, USA) followed by staining with a PE-coupled goat anti-human anti-Fc antibody (Jackson ImmunoResearch, West Grove, PA, USA). Data were acquired on a Fortessa FACS cytometer (BD Biosciences) and analyzed using FlowJo software (V10, Treestar, Ashland, OR).
2.3. CD22 density quantification in NALM6 cell lines
Fluorescence intensities from Quantibrite PE beads (BD Biosciences, Lot# 27504) were used to create a calibration curve with the average number of antibodies per bead listed by the manufacturer. The resulting line of best fit was used to calculate CD22 densities for each cell line.
2.4. In vitro cytotoxicity assay
The cytotoxic potential of the different CAR-T constructs against CD22+ leukemic cell lines was evaluated using Incucyte ® technology (Sartorius), as previously described [11]. Briefly, CAR-T cells were incubated at a 1:2 ratio with GFP+ NALM6 leukemia cells (25K/50K). NALM6 clones, characterized by different CD22 antigen densities, were utilized in co-culture experiments; CD22-overexpressing (CD22OE) and CD22-knockout (CD22KO) NALM6 have been described (Fry et al., 2018) and NALM6 expressing low levels of CD22 (CD22Low) were generated by FACS sorting on a FACSAria (BD Biosciences). Images, evaluating green fluorescence units per mm2 (GFU/mm2), were taken every 1 to 2 hours. 50,000 NALM6 cells were added per well to a 96-well plate and the wells were imaged to determine the initial GFU/mm2 prior to co-culture with CAR T-cells at a 1:2 effector : target ratio (E/T) in a total volume of 200 uL. GFU/mm2 was monitored by Phase Contrast Imaging every 1–2 hours. Killing Index was computed as the ratio of GFU/mm2 at each timepoint relative to the initial GFU/mm2.
2.5. Protein sample preparation
CD8α (UniProtKB# P01732) spanning amino acids F128 to D182 was sub-cloned into the pET15b vector (GenScript) and expressed in Escherichia coli strain BL21 (DE3) as a fusion protein with an N-terminal polyhistidine tag and a thrombin cleavage site. Cells were grown at 37°C to OD600 of 0.5 – 0.6 and protein expression induced at 17°C overnight by 0.4 mM IPTG. At the time of induction, zinc sulphate was added to a final concentration of 20 μM. The cells were frozen in liquid nitrogen and stored at −80°C for ~4 hours, followed by resuspension in buffer 1 (50 mM Tris at pH 7.5, 300 mM NaCl, 5 mM 2‑mercaptoethanol, 20 μM zinc sulphate) supplemented with EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics). Cells were lysed by sonication and spun down at 27,000 g for 30 min. The supernatant was incubated with pre-washed Ni-NTA agarose resin (Qiagen) for 1 hour. After extensive washing with buffer 1 and buffer 2 (50 mM Tris at pH 7.5, 50 mM NaCl, 5 mM 2‑mercaptoethanol, 20 μM zinc sulphate), CD8α (128–182) was separated from the His-tag and the resin by cleaving with thrombin (Millipore Sigma 605195) in buffer 2. Further purification was achieved by size exclusion chromatography on an FPLC ÄKTA pure system (GE Healthcare) using a HiLoad 16/600 Superdex 75 prep grade column (cytiva) in buffer 3 (20 mM NaPO4 at pH 6.5, 50 mM NaCl, 5 mM DTT, 20 μM zinc sulphate). 15N ammonium chloride (Cambridge Isotope Laboratories, Inc. NLM-467–1) and 13C glucose (Cambridge Isotope Laboratories, Inc. CLM-1396) were used for isotope labeling. All NMR samples were validated by mass spectrometry.
2.6. SDS-PAGE
Protein lysates were subjected to SDS-PAGE on 12% NuPAGE Bis-Tris gels (Thermo Fisher Scientific NP0342) with MOPS SDS running buffer (Thermo Fisher Scientific NP0001) and visualized by Coomasie staining.
2.7. Electrospray ionization mass spectrometry
Mass spectrometry was performed with CD8α H (10 μM) in buffer 3 with 10% acetonitrile on a 6100 Series Quadrupole LC mass spectrometer (Agilent Technologies, Inc.), equipped with an electrospray source and operated in the positive ion mode. Data acquisition and analyses were performed using OpenLAB CDS ChemStation Edition C.01.05 (Agilent Technologies, Inc.).
2.8. SEC-MALS
SEC-MALS data were collected by an Ultimate 3000 HPLC (Thermo Scientific) in-line with an Ultimate 3000 UV detector (Thermo Scientific), miniDawn MALS detector, and Optilab refractive index detector (Wyatt Technology). The data were collected following in-line fractionation with a WTC-010S5 (7.8 × 300 mm) 100 Å pore size SEC analytical column (Wyatt Technology), pre-equilibrated in 20 mM NaPO4 at pH 6.5, 50 mM NaCl, 20 μM zinc sulphate, with or without 5 mM DTT, running at a flow rate of 0.4 mL/min. 50 μL of BSA (30 μM, Thermo Fisher Scientific 23209), ubiquitin (500 μM, R&D systems U-100H-10M), or CD8α H (500 μM, either purified with or without DTT) was injected onto the column. Experiments were performed at 25°C. ASTRA software (version 8.0.2.5) was used for data collection and analyses.
2.9. NMR samples and experiments
Two NMR samples were prepared, namely 0.5 mM 15N, 13C-labeled CD8α H (sample 1) and 0.5 mM 15N-labeled CD8α H (sample 2). 2D 1H-15N HSQC and 1H-13C HSQC spectra and 3D HNCACB/CBCA(CO)NH, HNCO/HN(CA)CO, HACAN, (HB)CBCA(CO)N(CA)HA, 15N TOCSY-HSQC, HCCH-TOCSY, and 13C edited NOESY-HSQC spectra (250 ms mixing time) were recorded on sample 1. To optimize the mixing time of the 13C edited NOESY-HSQC spectrum, 2D 1H-1H plans with varying mixing time from 80 to 300 ms were recorded on sample 1. 2D 1H-15N HSQC and 15N-edited NOESY-HSQC (120 ms mixing time) spectra were recorded on sample 2. The EXSY experiment was recorded on sample 2 by monitoring the transfer of heteronuclear NZ magnetization during mixing times ranging from 10 to 100 ms. The conformational interconversion rate (Kconf) was analyzed from the EXSY data as described in [40–42] by using composite peak ratios of intensities of the auto-peaks and cross-peaks and fitting to equation 1 as a function of the mixing time t.
| (1) |
For two states X and Y, the intensities of cross-peaks originating on state Y and detected on state X are denoted aXY(t), whereases those originating on state X and detected on state Y are denoted aYX(t). aXX(t) and aYY(t) represent the intensities of the auto-peaks.
All NMR experiments were conducted in buffer 4 (20 mM NaPO4 at pH 6.5, 50 mM NaCl, 5 mM DTT, 20 μM zinc sulphate, 1 mM pefabloc, 0.1% NaN3, and 5% 2H2O / 95% 1H2O), except for 2D 1H-13C HSQC, 3D HCCH-TOCSY, and 13C-edited NOESY-HSQC experiments, which were acquired on samples dissolved in 2H2O. All NMR experiments were conducted at 25°C, except for 2D 1H-15N HSQC, 3D 15N TOCSY-HSQC, 15N-edited NOESY-HSQC spectra, and the EXSY experiment, which were acquired at 10°C. Spectra were recorded on Bruker AvanceIII 700, 800, or 850 MHz spectrometers equipped with cryogenically cooled probes.
All NMR data processing was performed using NMRpipe [43] and spectra were visualized and analyzed with XEASY [44]. Chemical shift index (CSI) values for CD8α Cα and C’ atoms were generated by comparison to random coil values of the same amino acid type and in the case of proline isomer state [45, 46] with sequence-dependent correction factors [47].
2.10. CD experiment
Far-UV range CD spectra (260–190 nm) of CD8α H (10 μM) were recorded on a Jasco J-1500 CD spectrometer using a quartz cuvette with 1.0 mm path length and temperature controlled at 25 ± 0.1°C. Buffer 3 was used as a control. All spectra were collected continuously at a scan speed of 20 nm/min and averaged over accumulation of three spectra. The buffer spectrum was subtracted from the protein spectra during data analyses. Molar ellipticity θ (in deg cm2 dmol−1) was calculated from the measured machine units m° in millidegrees at wavelength λ using Equation 2.
| (Eq. 2) |
C is the concentration of the sample in mol L−1 and L is the path length of the cell (cm). Secondary structure analysis was conducted with the program CDSSTR [48] by the DichroWeb server [49] using reference dataset 7 (190–240 nm) [48].
2.11. Model structure of the CD8α H in different states
To generate the model presented in Fig. 8A, the CD8α H was created and subsequently energy minimized with proline in the trans conformation in Schrödinger Maestro [50]. The cis proline isomers of states 2–4 were introduced by adjusting the dihedral O (Pro −1) - C’ (Pro −1) – N (Pro) – Cα (Pro) angle requiring a distance of 3.5 Å between the Hα atoms of proline and the preceding amino acid. Energy minimization was then repeated in Schrödinger Maestro.
Fig. 8.
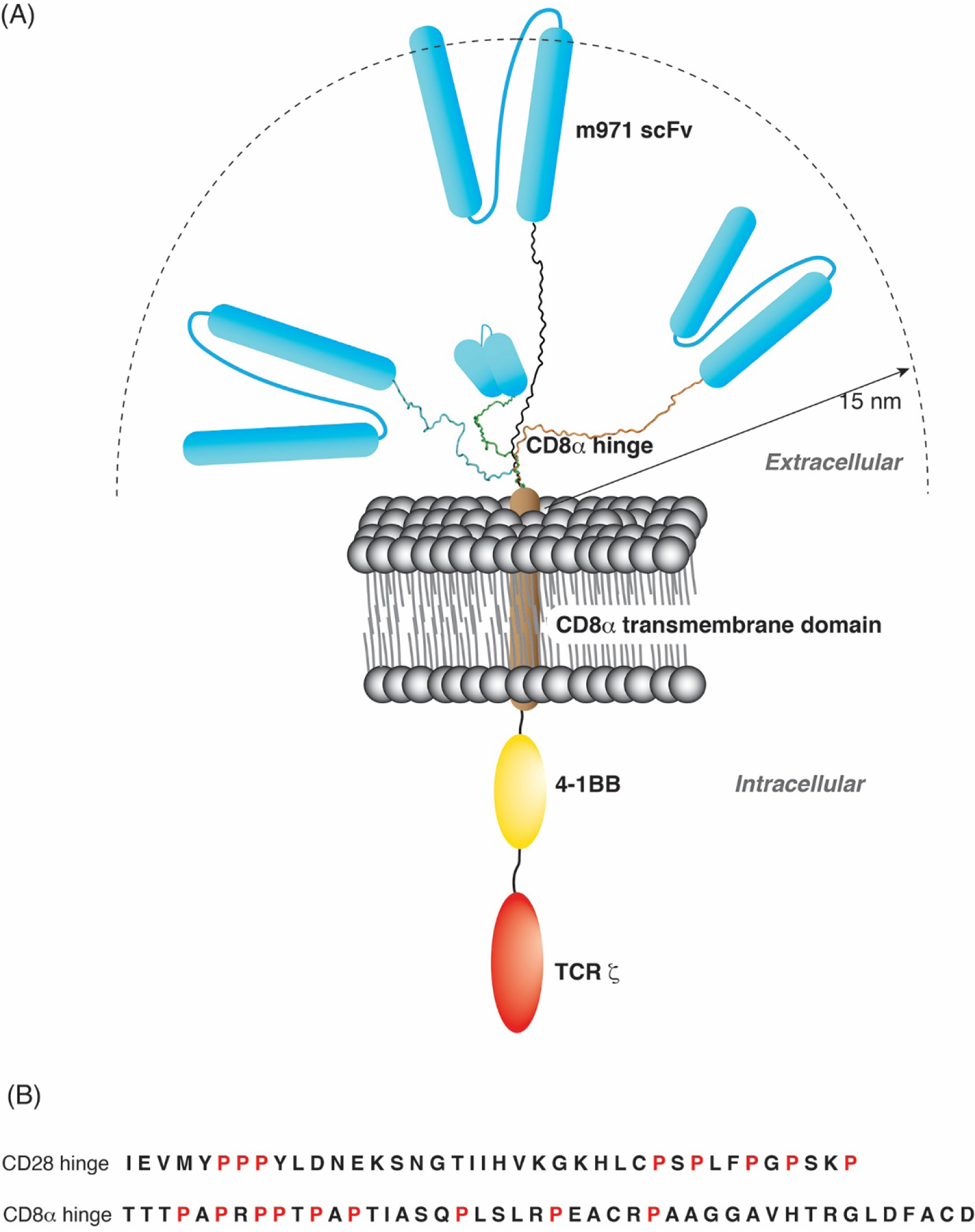
Schematic representation of CD22-CAR structure as a function of the CD8α and CD28 hinge domains. (A) Diagram of the CAR construct used in this study harboring the CD22-targeting m971 scFv (blue), CD8α TM (brown), 4–1BB costimulatory domain (yellow), as well as TCR ζ (red) as indicated. Four model structures (black, blue, green or orange) generated by Schrödinger maestro represent the CD8α H in the different states observed in NMR spectra. A distance of 15 nm is indicated as representing the intermembrane distance for T cell interaction with an antigen-presenting cell. (B) Amino acid sequence of the CD28 H and CD8α H under study with the prolines highlighted in red.
3. Results
3.1. A CD22-CAR harboring the CD8α H exhibits higher cytotoxicity against CD22-low leukemia cells than a CD28-H CAR
The cytotoxic potential of CD22-targeting CARs (m971 scFv) harboring either CD28 or CD8α H-TM domains were evaluated against CD22+ NALM6 leukemia. To eliminate differences due to intracellular signaling, all CARs were generated with a 4–1BB (CD137; TNFRS9) signaling domain (Fig. 1A) and all CARs were similarly expressed on transduced T cells (Fig. 1B). CD22-CART harboring either 28H-28TM or 8H-8TM were able to specifically eliminate NALM6 leukemia cells overexpressing CD22 (CD22OE; 25,980 molecules) with no killing detected against CD22-negative (CD22KO) Nalm6 (Fig. 1C). Cytotoxicity against CD22OE-Nalm6 was very efficient, with a killing index of 0.5 at approximately 24h following co-culture (Fig. 1D). However, the cytotoxic potential of CD22–28H-28TM against CD22Low-NALM6, harboring only 374 CD22 molecules (Fig. 1C), was significantly reduced as compared to the CD22–8H-8TM (Fig. 1D). Furthermore, this difference was specifically due to the hinge as a CD22-CAR harboring the 8H with a 28TM (8H-28TM) was as efficacious as an 8H-8TM construct (Fig. 1D). These data indicate the importance of an optimal H-TM when the targeted antigen is limiting. Notably though, hinge structure has yet to be elucidated and AlphaFold2 (AF2), the leading computational method for predicting protein structure [51, 52], predicts the CD8α H to be intrinsically disordered (Fig. 1E, left panel) but with low confidence (Fig. 1E, right panel).
Fig. 1.
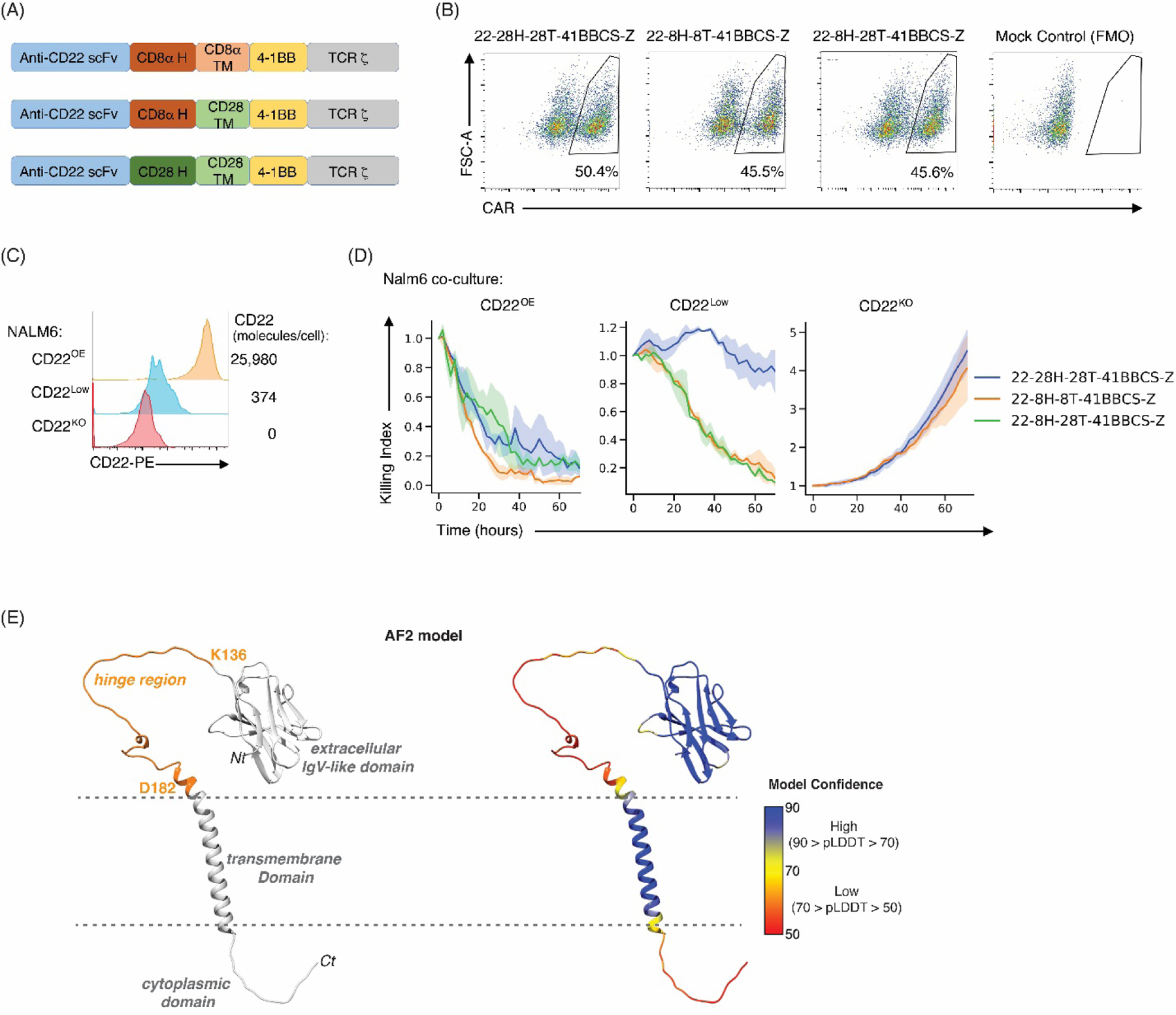
Generation of CD22 modular CAR constructs reveals the importance of the CD8α H region in cytotoxicity against CD22low leukemia. (A) Schematic representation of CD22 CAR constructs harboring the m971 scFv together with CD28 or CD8α hinge (H) and transmembrane (TM) domains together with 41BB costimulatory (CS) domain. (B) Human peripheral blood T cells were transduced with the indicated CAR constructs and cell surface CAR expression was evaluated using a CD22-Fc fusion protein. Representative dot plots are shown and percentages of CAR+ cells are indicated (1 of 4 biological replicates). (C) Representative histograms of CD22-overexpressing (CD22OE), CD22 low density (CD22Low), and CD22 knockout (CD22KO) NALM6 are shown (left). CD22 antigen density was quantified using Quantibrite-PE beads and numbers of cell surface CD22 molecules are indicated (right). (D) Ex vivo cytotoxicity was evaluated by co-culture of the indicated CAR-T with CD22OE, CD22low, or CD22KO NALM6 leukemia cells at a 1:2 ratio in an Incucyte assay. Killing index was evaluated as a function of time; solid lines show the average killing index and shaded areas are the standard error of the mean of biological replicates. Data are representative of 1 of 2 independent experiments. (E) Ribbon diagram of the AF2-predicted structure of CD8α illustrating the IgV-like extracellular domain, transmembrane helix, and cytoplasmic region in grey with the hinge region highlighted orange (left panel) or with coloration according to the AF2 per-residue confidence score (pLDDT) as indicated by the scale bar (right panel). UCSF Chimera [79] was used to make panel E.
3.2. The CD8α H exhibits intrinsic disorder
The CD8α H used in CARs spans F128 – D182 of homo sapiens CD8α (Fig. 2A). To study the structural properties of this hinge, we expressed and purified it from E. coli in frame with a His6 tag and thrombin cleavage site for purification by affinity chromatography. Further purification was achieved by size exclusion chromatography (SEC) with a Superdex 75 column on an FPLC system. During purification, the CD8α H eluted earlier than expected for its monomeric molecular weight (6,076 Da, with 4 amino acids remaining from the expression plasmid), but later than expected for a dimeric species, as indicated by comparison to 8.6 kDa ubiquitin and the 17 kDa hRpn13 Pru (Fig. 2B). To validate that the eluted protein was indeed CD8α, we performed liquid chromatography-mass spectrometry (LC-MS), which indicated a molecular weight of 6,074.76 Da (Fig. 2C). The early elution of CD8α H from the Superdex 75 column mimicked prior experiments performed for intrinsically disordered proteins, including hRpn10 RAZUL, Pup and SocB (Fig. 2B) [53–55].
Fig. 2.
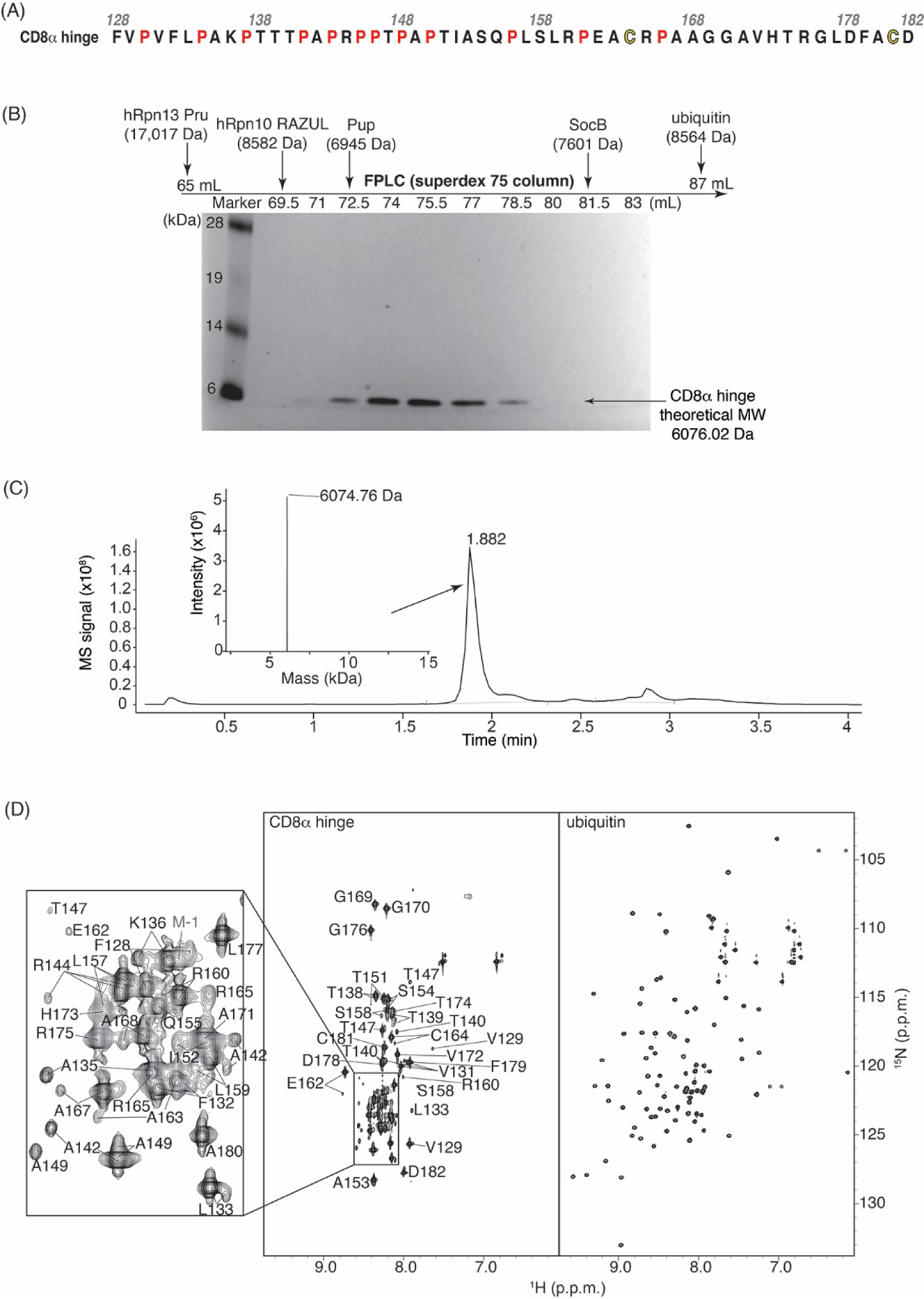
The CD8α H is intrinsically disordered. (A) Amino acid sequence of the CD8α H region under study with the prolines and cysteines highlighted in red and yellow respectively. (B) SDS-PAGE of fractions from SEC with a Superdex 75 column on an FPLC system for the CD8α H. A standard molecular weight marker is included in the left-most lane with labeling in kilodaltons (kDa). Elution volumes for other protein samples and their molecular weights are indicated above (chromatograms not shown). Like hRpn13 Pru, ubiquitin, and intrinsically disordered hRpn10 RAZUL, Pup, and SocB, CD8α elutes earlier than expected. (C) LC-MS analysis of purified CD8α H demonstrates its expected molecular weight. (D) Side-by-side view of 2D 1H, 15N HSQC spectra for the CD8α H (left) and ubiquitin (right, for comparison), with chemical shift assignments for the CD8α H region. The CD8α H lacks the amide chemical shift dispersion characteristic of a folded protein (as displayed for ubiquitin). All values are given in parts per million (p.p.m.) units.
To specifically test whether the CD8α H is intrinsically disordered, we used 2D NMR. A 1H, 15N HSQC experiment performed on 15N-labeled CD8α H was acquired to detect the amide groups. The resulting spectrum lacked chemical shift dispersion, with clustering of CD8α signals in the center of the spectrum (Fig. 2D, with ubiquitin for comparison on the right [56]. This phenomenon reflects lack of secondary structure, as we observed previously for other intrinsically disordered proteins [53, 54] and is consistent with the AF2 prediction (Fig. 1E).
3.3. The CD8α H is monomeric, but can form intermolecular disulfide bonds in the absence of reducing agent
The extracellular domain of CD8α forms a dimer (Fig. 3A, top right) and previous modeling has predicted that the hinge facilitates dimerization [28]. All AF2-Multimer predictions indicate two dimerization domains for CD8α; namely, the extracellular Ig domain in agreement with the experimentally determined domain structure (PDB: 2HP4, overall root-mean-square deviation between the Cα atoms of 0.7 Å) (Fig. 3A, left panel) and the TM, which is predicted to form a helical dimer. There are two cysteine residues in the CD8α H (Fig. 2A) that can, in principle, form disulfide bonds. The CD8α H is predicted to be intrinsically disordered and without intermolecular interactions except for these two cysteines (Fig. 3A, left panel). In some cases, AF2-Multimer predicted the TM helix to begin at either of these cysteines (most often at C181), initiated by their disulfide bonding.
Fig. 3.
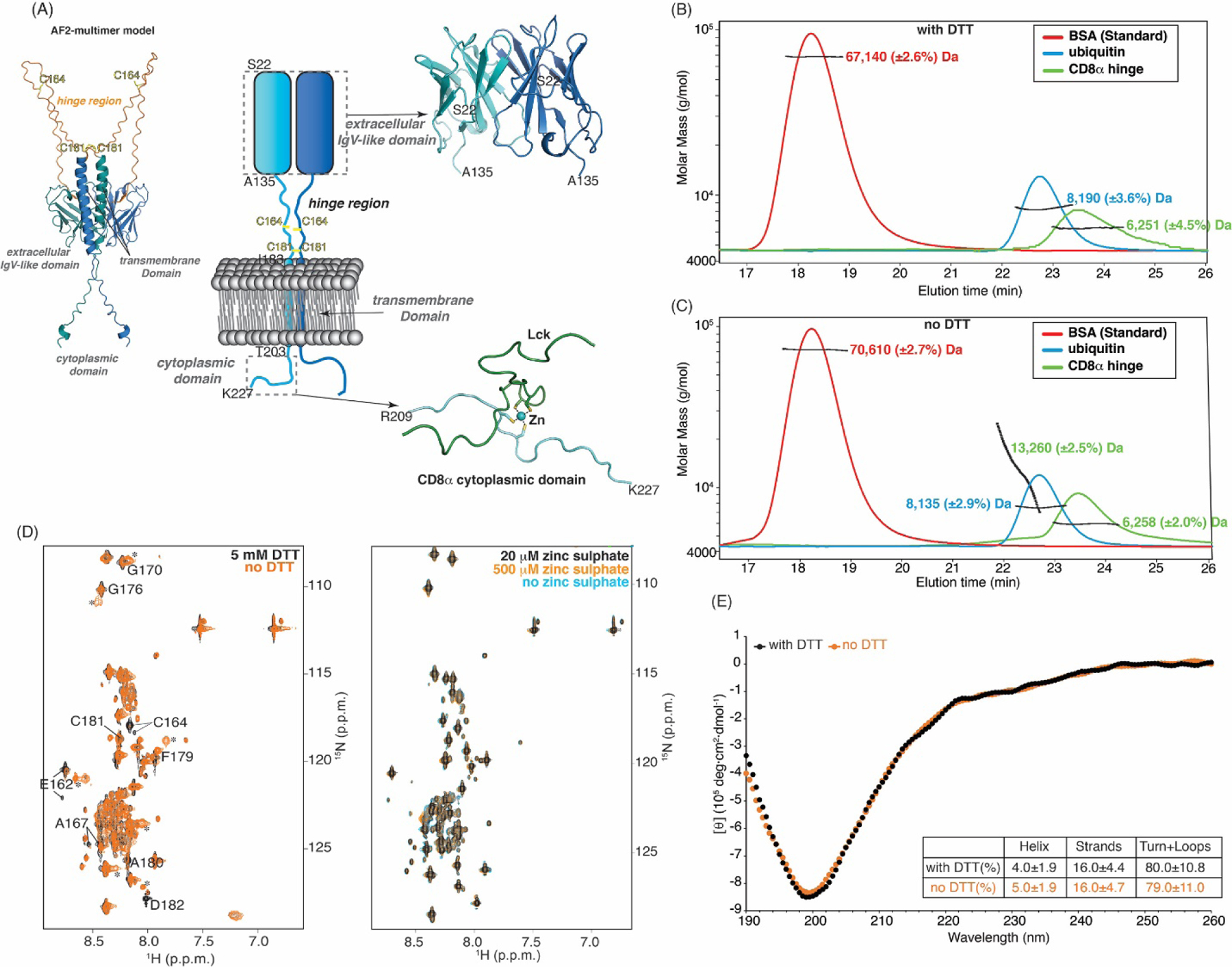
Long-range structure is not promoted in the CD8α H by disulfide bond formation or Zn2+. (A) Schematic representation (center) highlighting the dimerization of CD8α (shades of blue) as mediated through its extracellular IgV-like domain and ribbon diagram depicting Zn2+ (blue sphere) as a molecular glue for Lck (green) interaction with the CD8α cytoplasmic domain (bottom right, PDB:1Q69). Right top: ribbon diagram of the CD8α extracellular IgV-like domain dimer (light blue and dark blue, PDB: 2HP4). Left top: AF2-Multimer-predicted structure of the CD8α dimer with the hinge in orange. C164 and C181 in the CD8α H are labeled in yellow. (B, C) SEC-MALS analysis of bovine serum albumin (BSA, red, as a control, expected MW of 66.5 kDa), ubiquitin (blue, expected MW of 8564.7 Da), and CD8α H (green, expected MW of 6,076 Da) by using a 100 Å pore size SEC analytical column with (B) and no (C) DTT. MALS is plotted against elution time and determined molecular weights are provided. (D) 1H, 15N HSQC spectra of 15N-labeled CD8α H with 5 mM (black) or no (orange) DTT in buffer 4 (left panel) or with 20 μM (black), 500 μM (orange), or no zinc sulphate (blue) in buffer 4 containing DTT (right panel). Signals that shift or appear in the spectrum recorded without DTT are indicated with labels or asterisks; the spectrum without DTT is not assigned. (E) CD spectra of the CD8α H with (black) and without (orange) DTT in the far-UV region spanning from 190 to 260 nm at 25°C, with a table listing DichroWeb-predicted percentage of helix, strand, and unstructured regions. In (D) all values are given in p.p.m. units. Pymol (PyMOL Molecular Graphics System, http://www.pymol.org) was used to make Fig. A.
To experimentally probe the oligomerization state of the CD8α H, we used SEC-MALS (Wyatt Technology). As NMR experiments are performed at high concentrations (0.5 mM) compared to cellular levels, the experiments described in Figure 2 were performed in the presence of the dithiothreitol reducing agent (DTT), as routinely done [57]. We performed the SEC-MALS experiments both with and without DTT. With the smaller 100 Å pore size of the analytical SEC, the CD8α H eluted after 67 kDa BSA and 8.6 kDa ubiquitin and was found by MALS to have a molecular weight of 6,251 Da when DTT was present in the buffer (Fig. 3B). No signal was observed at higher molecular weights, indicating that no CD8α H oligomers were present in the sample with DTT. In the absence of DTT, molecular weights of the CD8α H were detected at 6,258 Da and 13,260 Da, reflective of a dimer (Fig. 3C). These data demonstrate that the CD8α H can form intermolecular disulfide bonds when its cysteines are not reduced, consistent with many of the AF2-Multimer models (Fig. 3A).
To test whether the presence of disulfide bonds in the CD8α H promotes structure, we used NMR and circular dichroism (CD). In support of the SEC-MALS data, the amide signals from the two cysteines (C164 and C181) and neighboring residues shift slightly depending on the presence of DTT in a 1H, 15N HSQC spectrum acquired on 15N labeled CD8α H. For example, new signals (marked by asterisks) were observed that appeared to correspond to E162, G170, G176, A180, and D182. However, the other signals were not affected by these changes (Fig. 3D, left panel). Thus, cysteine disulfide bonding causes spectral changes only for nearby amino acids, indicating that long-range structure is not induced. Similarly, CD spectroscopy, which informs on secondary structure, indicated intrinsic disorder both with and without DTT, with negative ellipticity observed near 200 nm and low ellipticity at 190 and 222 nm (Fig. 3E), similar to the intrinsically disordered proteins Pup and SocB [53, 54]. Furthermore, analysis of the CD spectrum by DichroWeb [58, 59] predicted 80% of the residues in the CD8α H to be unstructured, irrespective of the presence of DTT (Fig. 3E). The AF2-Multimer models of a full protein highlight the location of C181 proximal to the membrane where the TM helical dimer may promote its intermolecular disulfide bonding (Fig. 3A). Some models also show disulfide bridges at C164 (not shown). The CD data indicate that even in conditions with the cysteines are engaged in disulfide bonds, helicity is not induced in the CD8α H.
The C-terminal cytoplasmic tail of CD8α (spanning R209 – K227 in homo sapiens) and the N-terminus of the Src-family tyrosine kinase Lck form a folded heterodimeric structure with Zn2+ acting as a molecular glue (Fig. 3A, right bottom panel) [60]. Zinc sulphate (20 μM) was therefore included in the buffers throughout purification and added at induction, as done for the Zn-binding protein Put3 [61], unless otherwise noted. We did though directly test whether the presence of Zn2+ impacts the structure of the CD8α H. We acquired 1H, 15N HSQC spectra of CD8α H samples in the presence or absence of zinc sulphate present (20 μM and 500 μM, Fig. 3D, right panel). All three resulting spectra superimposed with no changes. This comparison revealed the CD8α H to be insensitive to Zn2+.
3.4. Four conformational states are observed for CD8α H residues
A greater than expected number of amide signals appeared in the 1H, 15N HSQC spectrum of 15N labeled CD8α H (Fig. 2D), suggesting that a subset of CD8α residues undergo chemical exchange between multiple states at a time scale slow enough for observation by NMR, as observed in other proteins with dynamic exchange between conformation states [62]. To assign the signals of the 2D spectrum to amide groups of CD8α, we acquired 3D 1H, 13C, 15N experiments that correlate sequential amino acids, as described in the Materials and Methods. The CD8α H is proline-rich (12 of 55 residues are prolines, Fig. 2A) and we therefore also used a HACAN spectrum [63] for assignment (Fig. 4A). Similar to the 2D 1H, 15N HSQC experiments (Fig. 2D), these 3D experiments indicated multiple signals for Cα (Fig. 4A and Fig. 4B, black signals in HNCACB and orange signals in CBCACONH) and Cβ (Fig. 4B, blue signals in HNCACB and orange signals in CBCACONH). Three distinct signals were observed for the V131, A135, T138, T147, A149, T151, L157 or E162 amide groups (Fig. 4A and 4B), and four signals were observed for A142 and R144 (Fig. 4B).
Fig. 4.
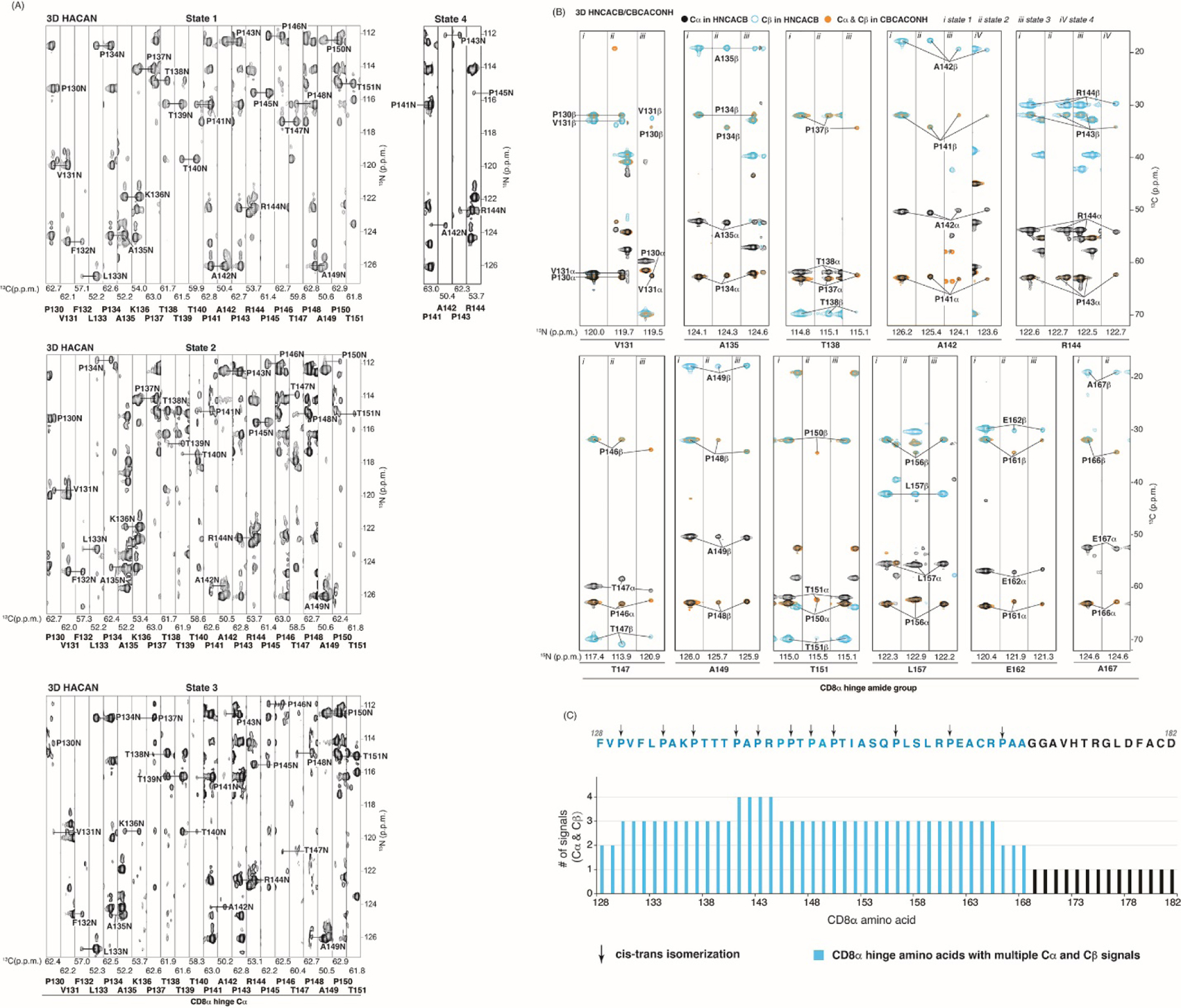
Up to four distinct states are observable in the CD8α H. (A) Strip plot series for CD8α H residues P130 – T151 generated by using 2D 1H, 15N planes of a 3D HACAN spectrum acquired at 850 MHz. Labels inside and below the strips correspond to N and Cα atoms respectively. For state 4, only strips P141 – R144 are displayed as all others are identical to state 1. (B) Strip plots of 2D 1H, 13C planes of a 3D HNCACB or CBCACONH spectrum for selected residues with greater than one set of resonances displaying Cα and Cβ signals in black and blue (HNACAB) or orange (CBCACONH) respectively. Labels within the strips correspond to carbon atoms as indicated with N atoms assigned below. The spectra of panels (A) and (B) were acquired with 0.5 mM 15N, 13C labeled CD8α H and plotted in p.p.m. units. (C) The number of Cα and Cβ signals observed for the CD8α H plotted across the sequence with those amino acids showing multiple Cα and Cβ signals highlighted in blue. Proline residues displaying signals for both cis and trans isomerization states are indicated with black arrows.
For each amino acid, the number of signals observed for the Cα and Cβ atoms was identical, and four signals corresponded to P141, A142, P143, and R144. Plotting these values across the sequence highlighted the signal multiplicity that spans from F128 to A168 of the CD8α H (Fig. 4C). In contrast, multiple signals were not observed for amino acids G169 – D182 (Fig. 4C). Thus, 3D spectra could be used to establish sequential amino acid assignments across the CD8α H sequence for each of the four states (Fig. 4A-B).
3.5. Prolines in the CD8α H exhibit trans and cis isomerization states
The CD8α H proline residues are confined to the region that undergoes chemical exchange (Fig. 4C). We therefore hypothesized that the prolines undergo cis-trans isomerization. Chemical shift values of proline Cβ and Cγ atoms inform on the cis versus trans isomer state [64]. All proline residues displayed multiplicity of signals that included chemical shifts reflective of the trans state, but 11 prolines also showed one set of signals indicative of a cis state (Table 1 and black arrows in Fig. 4C). We acquired a 13C-dispersed NOESY spectrum to directly evaluate the isomer state of the prolines with an optimized mixing time of 250 ms; this mixing time was chosen by comparing the signals in 2D 1H-1H planes acquired with different mixing times (Fig. 5A). NOEs from the proline Hα or Hδ atoms to the preceding residue’s Hα atom indicate cis or trans isomeric states respectively (Fig. 5B). We observed NOEs between proline Hδ and the preceding amino acid’s Hα for all prolines but P146, indicating trans isomerization (Fig. 5C). The chemical shift values of Cα and Hα for P146 are very similar to P145, causing the intra-residue NOE interactions to overlap with any possible inter-residue interactions, thus excluding P146 from this analysis.
Table 1.
Chemical shift values (p.p.m.) of the CD8α H proline carbonyl (C’), Cα, Cβ, Cγ, and N signals with State 1, 2, 3 and 4 indicated in black, blue, green, and orange, respectively.
| C’ | Cα | Cβ | Cγ | N | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| trans | cis | trans | cis | trans | cis | trans | cis | trans | cis | |
| P130 | 176.3, 176.0 |
175.7 | 62.7, 62.7 |
62.4 | 32.1, 32.0 |
34.3 | 27.1, 26.9 |
24.7 | 115.3, 115.3 |
114.3 |
| P134 | 176.4, 176.2 |
175.7 | 62.6, 62.5 |
62.4 | 31.9, 32.0 |
34.4 | 27.2, 27.1 |
24.6 | 112.8, 112.7 |
111.8 |
| P137 | 177.1, 177.0 |
176.2 | 63.0, 63.0 |
62.6 | 32.1, 32.2 |
34.3 | 27.1, 27.2 |
24.5 | 114.2, 114.2 |
112.7 |
| P141 | 176.2, 175.4, 175.1 |
175.5 | 62.8, 63.0, 63.0 |
62.6 | 32.0, 32.2, 32.1 |
34.4 | 27.0, 27.1, 27.1 |
24.7 | 116.3, 116.3, 116.3 |
114.9 |
| P143 | 176.6, 176.6, 176.6 |
175.9 | 62.7, 62.8, 62.8 |
62.3 | 32.0, 32.0, 32.0 |
34.3 | 26.9, 27.1, 26.9 |
24.5 | 112.5, 112.5, 112.4 |
112.0 |
| P145 | 61.4, 61.4, 62.2 |
30.7, 30.7, 30.4 |
27.2, 27.1, 27.2 |
115.6, 115.6, 115.5 |
||||||
| P146 | 176.7, 175.7 |
176.1 | 62.7, 63.0 |
62.5 | 31.9, 31.9 |
33.8 | 27.4, 26.8 |
24.5 | 112.2, 112.1 |
111.8 |
| P148 | 176.2, 176.1 |
175.6 | 62.8, 62.7 |
62.7 | 32.0, 31.9 |
34.2 | 26.9, 27.4 |
24.2 | 116.3, 115.0 |
114.8 |
| P150 | 177.0, 177.0 |
176.2 | 62.9, 62.9 |
62.4 | 32.0, 32.0 |
34.4 | 27.1, 27.2 |
24.8 | 112.4, 112.4 |
111.9 |
| P156 | 176.9, 176.8 |
176.2 | 63.1, 63.0 |
62.3 | 32.0, 32.0 |
34.3 | 27.1, 27.1 |
24.6 | 114.2, 114.2 |
112.7 |
| P161 | 177.3, 176.8 |
176.0 | 63.4, 63.1 |
62.5 | 32.0, 32.0 |
34.3 | 27.2, 27.3 |
24.7 | 114.3, 114.3 |
112.6, |
| P166 | 176.6 | 175.8 | 63.0 | 62.7 | 32.0 | 34.4 | 27.2 | 24.8 | 114.2 | 113.0 |
Fig. 5.
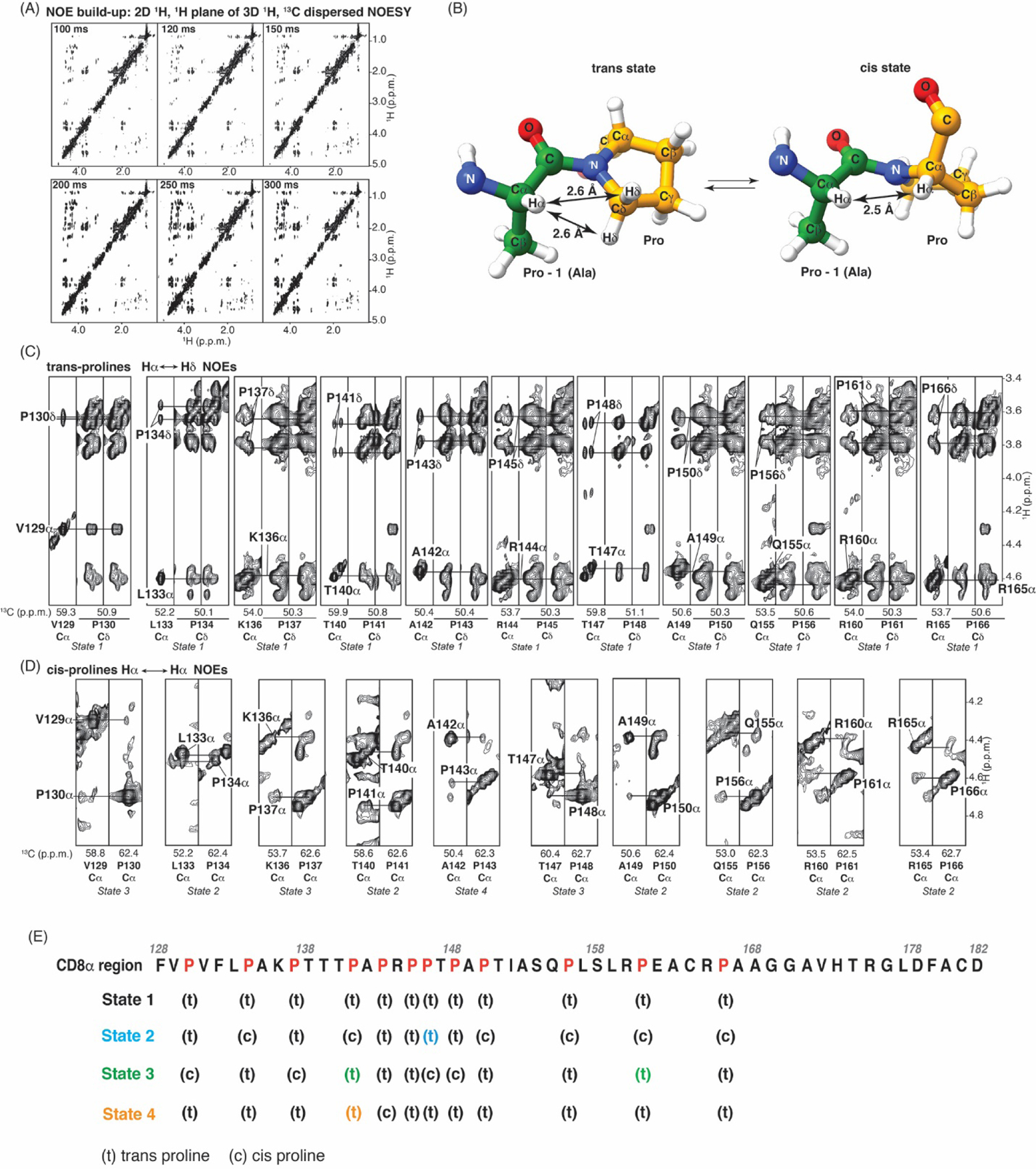
NOESY data confirm that the four CD8α H states are distinguishable by cis versus trans proline status. (A) 2D 1H, 1H planes of 1H, 13C dispersed NOESY experiment with mixing time of 100, 120, 150, 200, 250, or 300 ms. (B) Trans (left) and cis (right) proline (carbons in yellow) isomers with a preceding alanine (green) to demonstrate distances between the alanine Hα and proline Hδ (left) and Hα (right) atoms, as displayed and labeled. Oxygen, nitrogen, and hydrogen atoms are colored red, blue, and white, respectively. (C, D) Selected regions from a 1H, 13C-dispersed NOESY experiment with a mixing time of 250 ms illustrating NOE interactions between proline Hδ (C) and Hα (D) atoms and the preceding residue’s Hα; these NOEs indicate trans and cis isomerization states respectively. This spectrum was acquired with a sample dissolved in buffer 4 with 100% 2H2O. (E) Amino acid sequence of the CD8α H region with prolines indicated in red and assignments of cis and trans conformers for each of the four observable states. State 2, 3, or 4 prolines with trans configuration and C’ assignments that are different from state 1 are labeled in blue, green, or orange respectively.
In addition to the NOEs reflective of trans isomer states, NOE interactions between Hα atoms, which indicate the cis state, were observed for all prolines except P145 and P146 (Fig. 5D). NOEs between P145 and P146 Hα atoms could not be detected due to their similar values. However, the chemical shift values indicate P146 to be in the cis conformation for state 3. Hence, all prolines except for P145 can adopt either the cis or trans isomerization state. Moreover, multiple trans states were observable for all prolines except for P166. The cis/trans proline assignments were mapped onto the sequence of the four observable states in Fig. 5E.
3.6. Chemical shift values of the lesser populated states suggest local structural motifs
The CD data acquired on the CD8α H indicated a lack of global secondary structure (Fig. 3E). To provide insight into the structural characteristics of each of the four states, we generated chemical shift index (CSI) values by comparing the chemical shift values for Cα and C’ to random coil values of the same amino acid type, taking into account sequence-dependent correction factors [47]. No state displayed a sequential series of negative or positive values, which would reflect secondary structural elements (Fig. 6A-B). State 1 clearly indicated random coil values across the CD8α sequence. Fluctuations were observed however for the Cα values between the different states of V129, T140, T147, A149, E162, and T167 (Fig. 6A), all of which are located in a proline-rich region (Fig. 5E). We further evaluated this variance for the amide values and similarly detected large differences between the various states of V129, L133, T140, R144, T147, A149, E162, and A167 (Fig. 6C). We propose that the dispersed signals that appear in states 2–4 originate from regions with local structural motifs that are promoted by proline cis isomerization states.
Fig. 6.
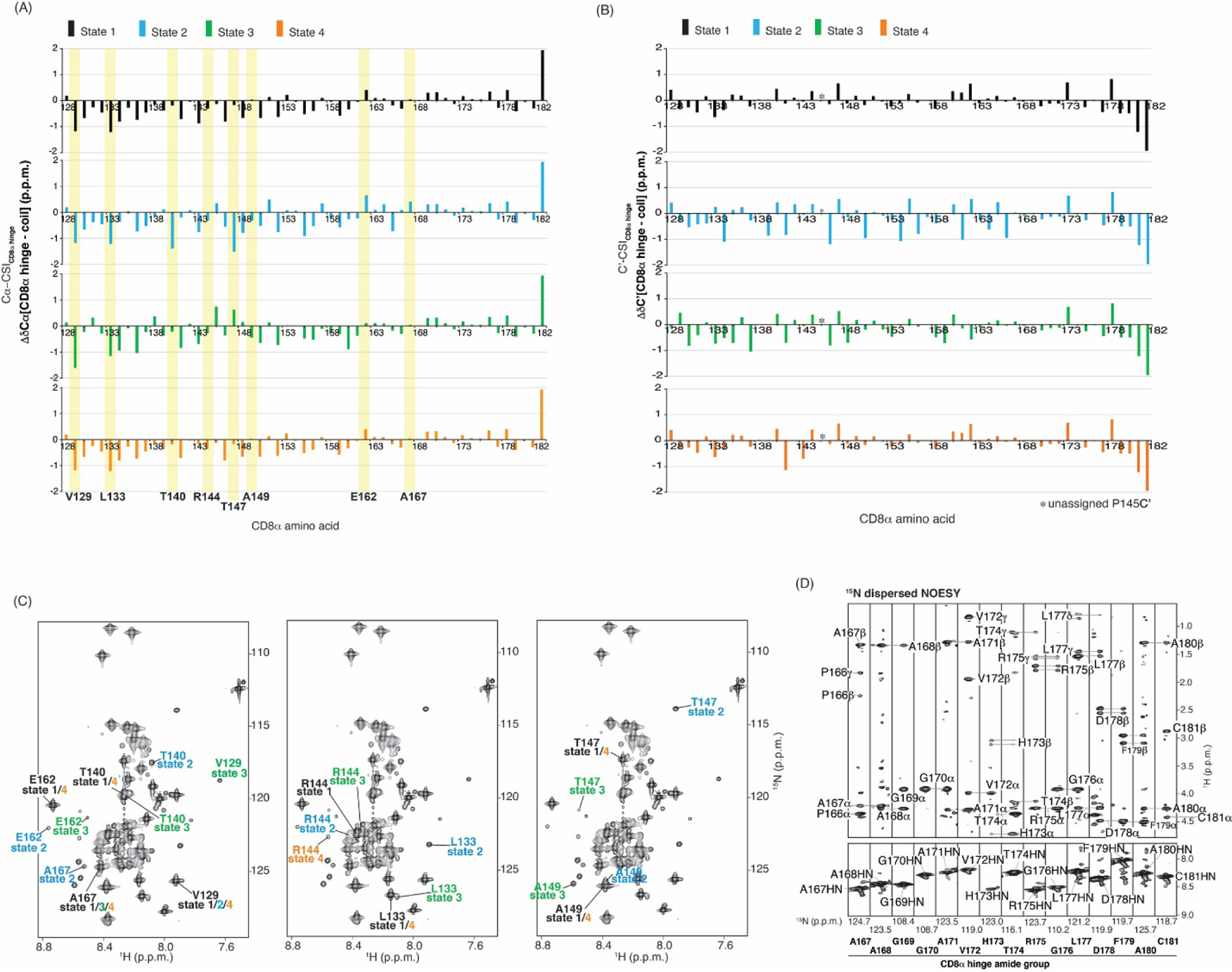
Lesser populated CD8α H states show spectral dispersion. (A, B) CSI profile for CD8α H Cα (A) or C’ (B) atoms for each of the four states. Yellow highlighting demarks V129, L133, T140, R144, T147, A149, E162 or A167. (C) 2D 1H, 15N HSQC spectra for the CD8α H showing assignments for V129, L133, T140, R144, T147, A149, E162 or A167 as indicated. (D) Regions of an 15N dispersed NOESY spectra acquired with 120 ms mixing time, labeling intra-residue and sequential NOE interactions. This spectrum was acquired with 0.5 mM 15N labeled CD8α H.
To further probe the structural features of the CD8α H, we acquired an 15N-dispersed NOESY experiment (120 ms mixing time) on 0.5 mM 15N-labeled CD8α H. Low abundance and corresponding poor signal-to-noise stymied the detection of intermolecular NOEs for states 2–4 and therefore this experiment was unable to inform on the structural features of these states. Consistent with the CSI data (Fig. 6A), state 1 showed only intra-residue and sequential NOE interactions (Fig. 6D), reflective of intrinsic disorder for this state.
We determined the relative abundance of states 1, 2, 3 and 4 to be 85%, 7%, 4%, and 4%, respectively. These values were obtained by measuring the Cα peak intensity of residues P141, A142, P143, and R144, which show clear signals for each of the four states in the HNCACB experiment (Fig. 4B). We expect that state 4 is distinguished from state 1 solely by cis-trans isomerization of P143, as all amino acids except for P141 – R144 have identical chemical shift values for these two states, indicating that structural effects are not propagated beyond the local neighboring sequence. In contrast, we note that P150, P156, P161, and P166 are either all cis or all trans in the observable states, suggesting a long-range correlation. In a longer construct, we expect that this correlation with a dimeric TM region may be further propagated by C164 and C181.
3.7. The conformational states of the CD8α H exchange dynamically
To directly test whether the four states observed within the CD8α H are in exchange, we acquired 1H, 15N, ZZ-exchange spectroscopy (EXSY) experiments with mixing times varying from 10 – 100 ms [65] at 25°C (not shown) and 10°C (Fig. 7A). Spectral crowding limited observation of EXSY exchange peaks (Fig. 2D and 7B), such as signals from different states of F128 or L159, or those will similar amide values, such as A135 or T151; these cases were therefore excluded from this analysis and labeled as “§” in Fig. 7A. Exchange peaks were observed for most amino acids with sufficient chemical shift dispersion (Fig. 7A), as illustrated for A149, S158, A163, and A167 (Fig. 7B and 7C). For two amino acids (V129 and T147), dispersion was sufficient but EXSY exchange peaks were not observed at neither 10°C nor 25°C. Moreover, exchanges peaks were only observed between state 1 and state 2 or 3.
Fig. 7.
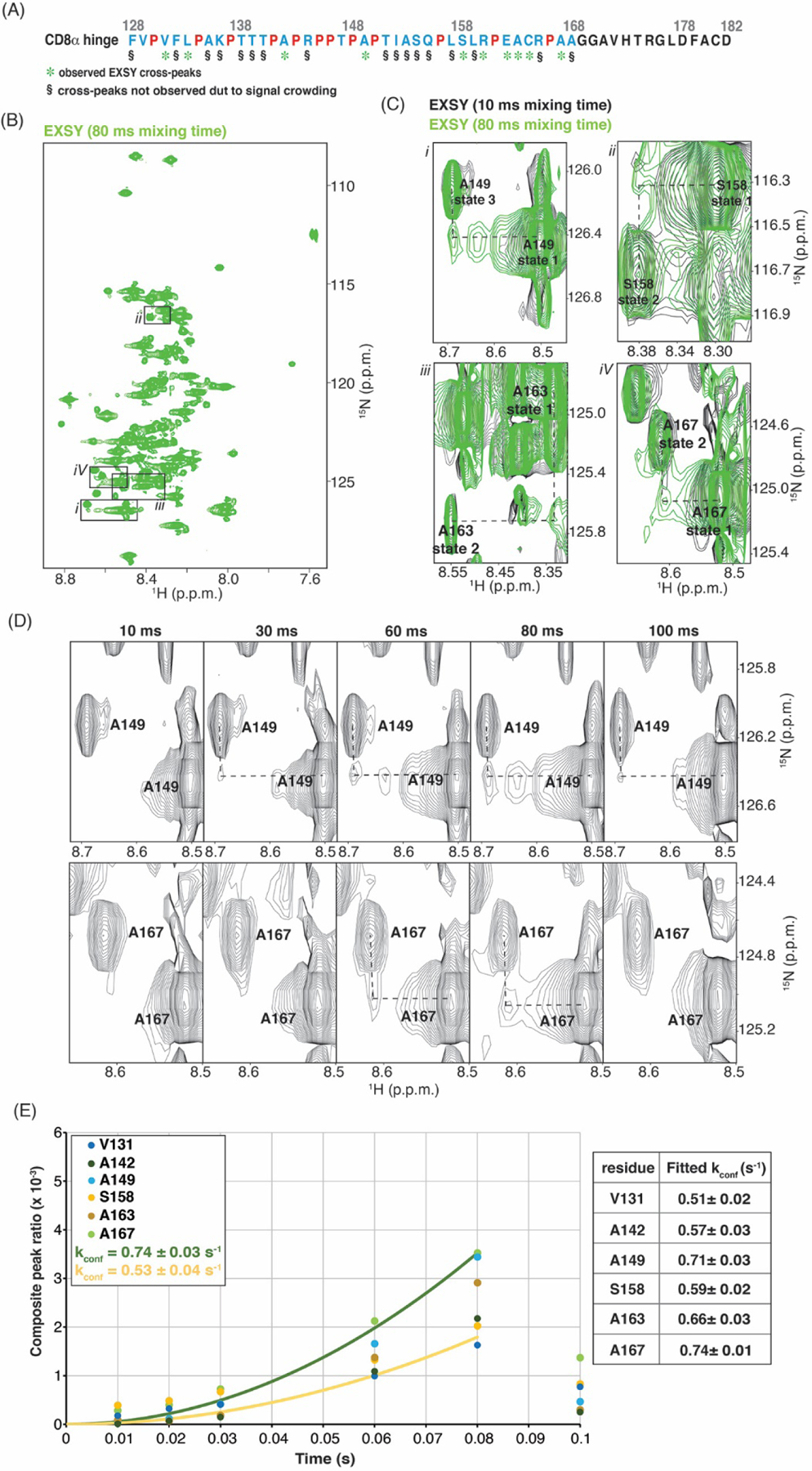
The CD8α H undergoes dynamic exchange. (A) Amino acid sequence of the CD8α H region as in Fig. 4C but with green asterisks below residues that show EXSY exchange signals. (B) EXSY spectrum of the CD8α H with 80 ms mixing time. (C) Enlarged regions of overlaid EXSY spectra with 10 ms (black) and 80 ms (green) mixing time demonstrating cross-peaks for A149, S158, A163 and A167. The enlarged regions are indicated by black boxes in (B). (D) Enlarged regions of EXSY spectra for A149 and A167 with mixing times of 10, 30, 60, 80 and 100 ms. (E) Exchange buildup plotted against time and fit to rate values for selected amino acids (left) with values provided in a table (right). In (B), (C) and (D), all values are given in parts per million (p.p.m.) units.
The exchange peak signals increased over time to 80 ms and then decayed by 100 ms (Fig. 7D). The build-up to 80 ms was fit to quantitatively determine a conformational interconversion rate (Kconf) between 0.53 – 0.74 s−1 for exchange between state 1 and state 2, based on V131, A142, S158, A163, and A167, and between state 1 and state 3, based on A149 (Fig. 7E). Altogether, these findings indicate that the CD8α H undergoes dynamic conformational exchange between different conformational states.
4. Discussion
Differences in the length, flexibility and composition of the hinge region can affect CAR expression, signaling, epitope recognition, and strength of activation outputs [66, 67]. We report here that the CD8α H region has four distinct conformational states of differing populations, with the most abundantly populated state being intrinsically disordered. Dispersed chemical shift values of certain amino acids in the NMR spectra suggest that local structural motifs are formed in the minor states.
We propose that the conformational exchange between the CD8α H states is driven by proline cis-trans isomerization. Supporting this model is the finding that exchange is absent in the C-terminal quartile of the sequence that lacks prolines. Proline generally adopts the trans isomeric state [68] and in State 1, which is most abundant and intrinsically disordered state, all prolines are all trans (Fig. 5E). A distinguishing feature of the other states is the presence of prolines that are in the cis conformation (Table 1). Cis isomeric states induce kinking in the chain, and we propose that this effect promotes local structural motifs. The cis/trans conformational state of certain prolines appear to be coordinated; for example, the four most C-terminally located prolines at positions P150, P156, P161, and P166 are either all cis or all trans. To determine whether the proline exchange is independent or coordinated, future studies with amino acid substitutions are planned.
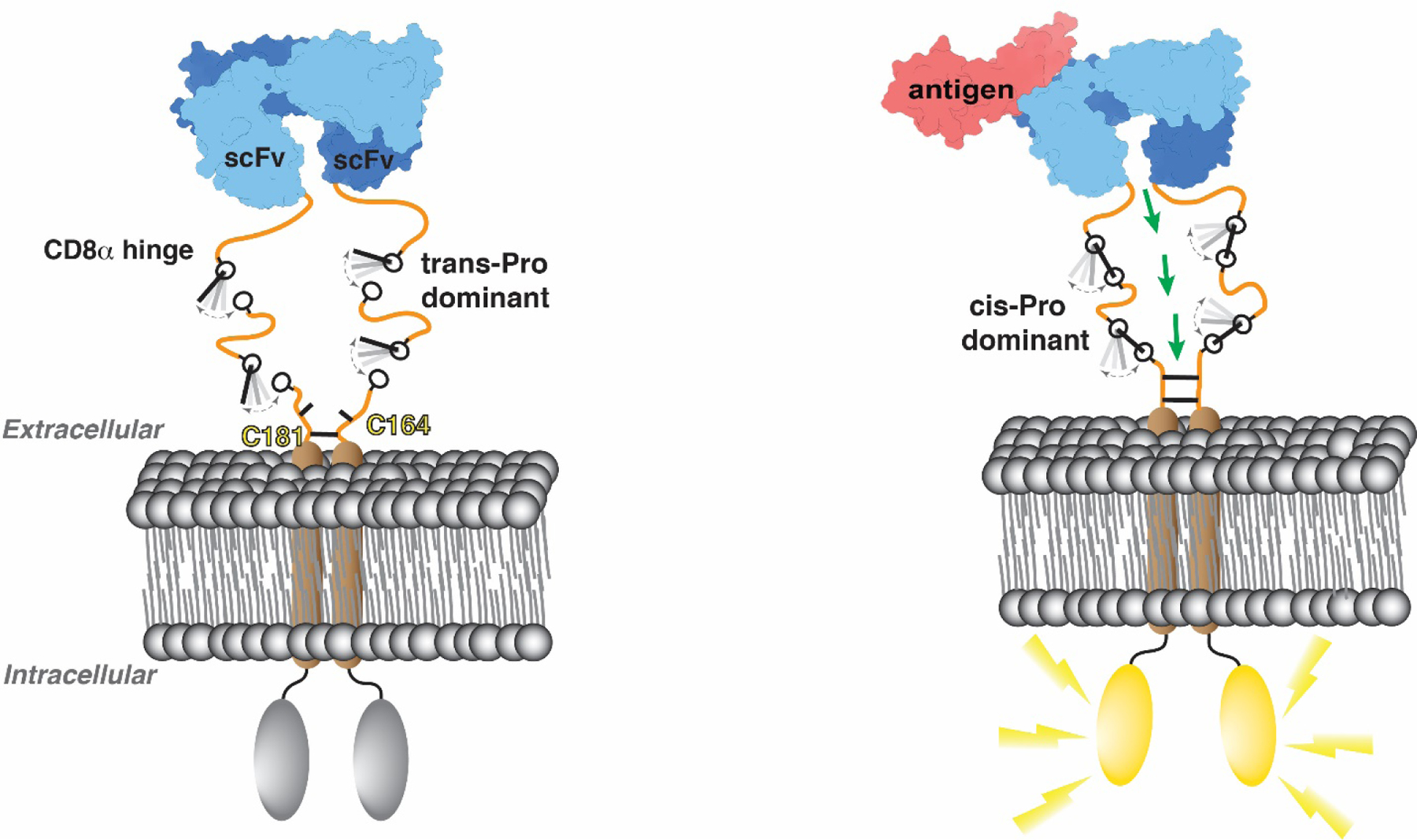
A T cell interacts with an antigen-presenting cell to form an immunological synapse with an intermembrane distance between the extended T cell receptor (TCR) and the peptide-major histocompatibility complex (MHC) complex of ~15 nm [69]; molecules with extracellular domains longer than this distance are sterically excluded. [69]. Proline cis-trans isomerization in the CD8α H appears to drive conformational exchange and CAR structural plasticity with cis proline states promoting local structural motifs that may act as switches to increase local order and perhaps thereby enhance allostery between the extracellular antigen binding domain and the intracellular signaling domain (Fig. 8A). This allostery may be amplified intermolecularly in full proteins by disulfide bridges at C164 and C181. We propose that the dynamic switching and the local structural features enabled by proline in combination with disulfide bridging between dimeric molecules promotes signaling in the low antigen context. This model requires future experiments, and a limitation of our study is that the isolated H domain may behave differently compared to the full-length CAR in the cell membrane setting. Interactions with the cell surface may stabilize certain local structures and/or alter exchange dynamics.
In the context of our study, which showed that a CD22-CAR with a CD8α H has more efficacy than a CD28 H against a CD22 low density leukemia (Fig. 1A-D), it is interesting to note that the CD28 H is shorter and contains less prolines (Fig. 8B). Three of the CD28 H prolines are located at the N-terminal end, neighbored by tyrosines, and the remaining five prolines are distributed at the C-terminal end (Fig. 8B). It is likely that the 3-residue proline sequence in CD28 also contains cis isomers, as neighboring aromatic residues are reported to promote cis proline isomers [70] by π-CH interactions [71]; this feature may make CD28 superior as a hinge in certain contexts and in future work, it will be important to evaluate the relative activity of a CD28 H as a function of different CAR transmembrane domains. We propose that proline residues in the CAR H regions as well as the presence of cis-promoting neighboring amino acids may be important for CAR activity.
Peptidyl prolyl cis/trans isomerases (PPlases) specifically enhance proline cis-trans isomerization [72] and act on CD8α during endogenous activities. The mitochondrial PPlase Cyclophilin D promotes CD8+ T cell-mediated response to viral infection [73]. Also, CD8+ T cell proliferation in response to infection with L. monocytogenes is impaired by loss of PPIase Pin1 [74], which catalyzes cis-trans interconversions of pSer/pThr-Pro peptide bonds [75]. As the CD8α H contains two Thr-Pro peptide bonds (Fig. 8B), it will be of interest to investigate whether CAR T-cell therapies would benefit from inclusion of a PPlase such as Cyclophilin D or Pin1. This is critical in the context of low antigen density tumor targets such as CD22 wherein antigen loss and downregulation result in a resistance to CD22 CART cells [76–78]. In conclusion, our study demonstrates structural plasticity in the CD8α H characterized by exchange on the millisecond time scale between a dominant intrinsically disordered state and ones with local structural features. This exchange and the local structure is promoted by cis-trans isomerization of dispersed prolines. Future studies aimed at testing the importance of these prolines in the CD8α H for cytotoxicity may lead to improved engineering of CARTs.
Highlights.
CD8α outperforms CD28 hinges in CD22-CARs at killing low antigen leukemia cells
CD8α hinge residues exchange dynamically between distinct states observable by NMR
The dominant CD8α hinge state is intrinsically disordered with only trans prolines
Sparsely populated states contain cis prolines and apparent local structural motifs
Prolines in the CD8α hinge undergo cis-trans isomerization
Acknowledgements
This work was funded by the Intramural Research Program through the Center for Cancer Research, National Cancer Institute, National Institutes of Health (ZIA BC011627 to K.J.W and ZIA BC 011923 to N.T.) and by federal funds from the National Cancer Institute, National Institutes of Health under contract 75N91019D00024 to H.M. We thank Angela Su for her assistance in generating Nalm6 cell lines, Janusz Koscielniak for maintenance of the NMR spectrometers, and the Biophysics Resource in the Center for Structural Biology, CCR for assistance with LC-MS and CD-spectroscopy. We are grateful to Dr. Tom Misteli for his important input in the initiation of this project. AF2 and AF2-Multimer modeling utilized the computational resources of the High-Performance Computing Biowulf cluster of the NIH (http://hpc.nih.gov). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Restifo NP, Dudley ME, Rosenberg SA, Adoptive immunotherapy for cancer: harnessing the T cell response, Nat Rev Immunol, 12 (2012) 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Eshhar Z, Waks T, Gross G, Schindler DG, Specific activation and targeting of cytotoxic lymphocytes through chimeric single chains consisting of antibody-binding domains and the gamma or zeta subunits of the immunoglobulin and T-cell receptors, Proc Natl Acad Sci U S A, 90 (1993) 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bollard CM, Tripic T, Cruz CR, Dotti G, Gottschalk S, Torrano V, Dakhova O, Carrum G, Ramos CA, Liu H, Wu MF, Marcogliese AN, Barese C, Zu Y, Lee DY, O’Connor O, Gee AP, Brenner MK, Heslop HE, Rooney CM, Tumor-Specific T-Cells Engineered to Overcome Tumor Immune Evasion Induce Clinical Responses in Patients With Relapsed Hodgkin Lymphoma, J Clin Oncol, 36 (2018) 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ramos CA, Grover NS, Beaven AW, Lulla PD, Wu MF, Ivanova A, Wang T, Shea TC, Rooney CM, Dittus C, Park SI, Gee AP, Eldridge PW, McKay KL, Mehta B, Cheng CJ, Buchanan FB, Grilley BJ, Morrison K, Brenner MK, Serody JS, Dotti G, Heslop HE, Savoldo B, Anti-CD30 CAR-T Cell Therapy in Relapsed and Refractory Hodgkin Lymphoma, J Clin Oncol, 38 (2020) 3794–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kalos M, Levine BL, Porter DL, Katz S, Grupp SA, Bagg A, June CH, T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia, Sci Transl Med, 3 (2011) 95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris MR, Stefanski HE, Myers GD, Qayed M, De Moerloose B, Hiramatsu H, Schlis K, Davis KL, Martin PL, Nemecek ER, Yanik GA, Peters C, Baruchel A, Boissel N, Mechinaud F, Balduzzi A, Krueger J, June CH, Levine BL, Wood P, Taran T, Leung M, Mueller KT, Zhang Y, Sen K, Lebwohl D, Pulsipher MA, Grupp SA, Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia, N Engl J Med, 378 (2018) 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gardner RA, Finney O, Annesley C, Brakke H, Summers C, Leger K, Bleakley M, Brown C, Mgebroff S, Kelly-Spratt KS, Hoglund V, Lindgren C, Oron AP, Li D, Riddell SR, Park JR, Jensen MC, Intent-to-treat leukemia remission by CD19 CAR T cells of defined formulation and dose in children and young adults, Blood, 129 (2017) 3322–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, Liedtke M, Rosenblatt J, Maus MV, Turka A, Lam LP, Morgan RA, Friedman K, Massaro M, Wang J, Russotti G, Yang Z, Campbell T, Hege K, Petrocca F, Quigley MT, Munshi N, Kochenderfer JN, Anti-BCMA CAR T-Cell Therapy bb2121 in Relapsed or Refractory Multiple Myeloma, N Engl J Med, 380 (2019) 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Roex G, Timmers M, Wouters K, Campillo-Davo D, Flumens D, Schroyens W, Chu Y, Berneman ZN, Lion E, Luo F, Anguille S, Safety and clinical efficacy of BCMA CAR-T-cell therapy in multiple myeloma, J Hematol Oncol, 13 (2020) 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Spiegel JY, Patel S, Muffly L, Hossain NM, Oak J, Baird JH, Frank MJ, Shiraz P, Sahaf B, Craig J, Iglesias M, Younes S, Natkunam Y, Ozawa MG, Yang E, Tamaresis J, Chinnasamy H, Ehlinger Z, Reynolds W, Lynn R, Rotiroti MC, Gkitsas N, Arai S, Johnston L, Lowsky R, Majzner RG, Meyer E, Negrin RS, Rezvani AR, Sidana S, Shizuru J, Weng WK, Mullins C, Jacob A, Kirsch I, Bazzano M, Zhou J, Mackay S, Bornheimer SJ, Schultz L, Ramakrishna S, Davis KL, Kong KA, Shah NN, Qin H, Fry T, Feldman S, Mackall CL, Miklos DB, CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: a phase 1 trial, Nat Med, 27 (2021) 1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, Wolters P, Martin S, Delbrook C, Yates B, Shalabi H, Fountaine TJ, Shern JF, Majzner RG, Stroncek DF, Sabatino M, Feng Y, Dimitrov DS, Zhang L, Nguyen S, Qin H, Dropulic B, Lee DW, Mackall CL, CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy, Nat Med, 24 (2018) 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].O’Leary MC, Lu X, Huang Y, Lin X, Mahmood I, Przepiorka D, Gavin D, Lee S, Liu K, George B, Bryan W, Theoret MR, Pazdur R, FDA Approval Summary: Tisagenlecleucel for Treatment of Patients with Relapsed or Refractory B-cell Precursor Acute Lymphoblastic Leukemia, Clin Cancer Res, 25 (2019) 1142–1146. [DOI] [PubMed] [Google Scholar]
- [13].Pulte ED, Vallejo J, Przepiorka D, Nie L, Farrell AT, Goldberg KB, McKee AE, Pazdur R, FDA Supplemental Approval: Blinatumomab for Treatment of Relapsed and Refractory Precursor B-Cell Acute Lymphoblastic Leukemia, Oncologist, 23 (2018) 1366–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rafei H, Kantarjian HM, Jabbour EJ, Recent advances in the treatment of acute lymphoblastic leukemia, Leuk Lymphoma, 60 (2019) 2606–2621. [DOI] [PubMed] [Google Scholar]
- [15].Inaba H, Pui CH, Immunotherapy in pediatric acute lymphoblastic leukemia, Cancer Metastasis Rev, 38 (2019) 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Porter DL, Levine BL, Kalos M, Bagg A, June CH, Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia, N Engl J Med, 365 (2011) 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liang A, Ye S, Li P, Huang J, Zhu S, Yao X, Zhou L, Xu Y, Zhu J, Zheng C, Chen S, Lan L, Lv X, Wei Y, Humphries M, Yao Y, Safety and efficacy of a novel anti-CD20 chimeric antigen receptor (CAR)-T cell therapy in relapsed/refractory (r/r) B-cell non-Hodgkin lymphoma (B-NHL) patients after failing CD19 CAR-T therapy, Journal of Clinical Oncology, 39 (2021) 2508–2508. [Google Scholar]
- [18].Shah NN, Johnson BD, Schneider D, Zhu F, Szabo A, Keever-Taylor CA, Krueger W, Worden AA, Kadan MJ, Yim S, Cunningham A, Hamadani M, Fenske TS, Dropulić B, Orentas R, Hari P, Bispecific anti-CD20, anti-CD19 CAR T cells for relapsed B cell malignancies: a phase 1 dose escalation and expansion trial, Nature Medicine, 26 (2020) 1569–1575. [DOI] [PubMed] [Google Scholar]
- [19].Dai H, Wu Z, Jia H, Tong C, Guo Y, Ti D, Han X, Liu Y, Zhang W, Wang C, Zhang Y, Chen M, Yang Q, Wang Y, Han W, Bispecific CAR-T cells targeting both CD19 and CD22 for therapy of adults with relapsed or refractory B cell acute lymphoblastic leukemia, Journal of Hematology & Oncology, 13 (2020) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cronk RJ, Zurko J, Shah NN, Bispecific Chimeric Antigen Receptor T Cell Therapy for B Cell Malignancies and Multiple Myeloma, Cancers, 12 (2020) 2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wagner J, Wickman E, DeRenzo C, Gottschalk S, CAR T Cell Therapy for Solid Tumors: Bright Future or Dark Reality?, Mol Ther, 28 (2020) 2320–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yong CSM, Dardalhon V, Devaud C, Taylor N, Darcy PK, Kershaw MH, CAR T-cell therapy of solid tumors, Immunol Cell Biol, 95 (2017) 356–363. [DOI] [PubMed] [Google Scholar]
- [23].Sievers NM, Dörrie J, Schaft N, CARs: Beyond T Cells and T Cell-Derived Signaling Domains, International Journal of Molecular Sciences, 21 (2020) 3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Holzinger A, Abken H, CAR T Cells: A Snapshot on the Growing Options to Design a CAR, HemaSphere, 3 (2019) e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Abate-Daga D, Davila ML, CAR models: next-generation CAR modifications for enhanced T-cell function, Molecular Therapy - Oncolytics, 3 (2016) 16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fujiwara K, Tsunei A, Kusabuka H, Ogaki E, Tachibana M, Okada N, Hinge and Transmembrane Domains of Chimeric Antigen Receptor Regulate Receptor Expression and Signaling Threshold, Cells, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Alabanza L, Pegues M, Geldres C, Shi V, Wiltzius JJW, Sievers SA, Yang S, Kochenderfer JN, Function of Novel Anti-CD19 Chimeric Antigen Receptors with Human Variable Regions Is Affected by Hinge and Transmembrane Domains, Mol Ther, 25 (2017) 2452–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Muller YD, Nguyen DP, Ferreira LMR, Ho P, Raffin C, Valencia RVB, Congrave-Wilson Z, Roth TL, Eyquem J, Van Gool F, Marson A, Perez L, Wells JA, Bluestone JA, Tang Q, The CD28-Transmembrane Domain Mediates Chimeric Antigen Receptor Heterodimerization With CD28, Front Immunol, 12 (2021) 639818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ramello MC, Benzaïd I, Kuenzi BM, Lienlaf-Moreno M, Kandell WM, Santiago DN, Pabón-Saldaña M, Darville L, Fang B, Rix U, Yoder S, Berglund A, Koomen JM, Haura EB, Abate-Daga D, An immunoproteomic approach to characterize the CAR interactome and signalosome, Sci Signal, 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Majzner RG, Rietberg SP, Sotillo E, Dong R, Vachharajani VT, Labanieh L, Myklebust JH, Kadapakkam M, Weber EW, Tousley AM, Richards RM, Heitzeneder S, Nguyen SM, Wiebking V, Theruvath J, Lynn RC, Xu P, Dunn AR, Vale RD, Mackall CL, Tuning the Antigen Density Requirement for CAR T-cell Activity, Cancer Discov, 10 (2020) 702–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Brudno JN, Lam N, Vanasse D, Shen YW, Rose JJ, Rossi J, Xue A, Bot A, Scholler N, Mikkilineni L, Roschewski M, Dean R, Cachau R, Youkharibache P, Patel R, Hansen B, Stroncek DF, Rosenberg SA, Gress RE, Kochenderfer JN, Safety and feasibility of anti-CD19 CAR T cells with fully human binding domains in patients with B-cell lymphoma, Nat Med, 26 (2020) 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW, The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor, Cell, 127 (2006) 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Call ME, Wucherpfennig KW, Chou JJ, The structural basis for intramembrane assembly of an activating immunoreceptor complex, Nat Immunol, 11 (2010) 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mineev KS, Goncharuk SA, Arseniev AS, Toll-like receptor 3 transmembrane domain is able to perform various homotypic interactions: an NMR structural study, FEBS Lett, 588 (2014) 3802–3807. [DOI] [PubMed] [Google Scholar]
- [35].Wittlich M, Thiagarajan P, Koenig BW, Hartmann R, Willbold D, NMR structure of the transmembrane and cytoplasmic domains of human CD4 in micelles, Biochimica et biophysica acta, 1798 (2010) 122–127. [DOI] [PubMed] [Google Scholar]
- [36].Haso W, Lee DW, Shah NN, Stetler-Stevenson M, Yuan CM, Pastan IH, Dimitrov DS, Morgan RA, FitzGerald DJ, Barrett DM, Wayne AS, Mackall CL, Orentas RJ, Anti-CD22– chimeric antigen receptors targeting B-cell precursor acute lymphoblastic leukemia, Blood, 121 (2013) 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Carpenito C, Milone MC, Hassan R, Simonet JC, Lakhal M, Suhoski MM, Varela-Rohena A, Haines KM, Heitjan DF, Albelda SM, Carroll RG, Riley JL, Pastan I, June CH, Control of large, established tumor xenografts with genetically retargeted human T cells containing CD28 and CD137 domains, Proc Natl Acad Sci U S A, 106 (2009) 3360–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kucera R, Cantor E, Golden Gate Assembly Protocol for Using NEB Golden Gate Assembly Mix, in, 2018.
- [39].Qin H, Ramakrishna S, Nguyen S, Fountaine TJ, Ponduri A, Stetler-Stevenson M, Yuan CM, Haso W, Shern JF, Shah NN, Fry TJ, Preclinical Development of Bivalent Chimeric Antigen Receptors Targeting Both CD19 and CD22, Molecular Therapy - Oncolytics, 11 (2018) 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wagner G, Bodenhausen G, Mueller N, Rance M, Soerensen OW, Ernst RR, Wuethrich K, Exchange of two-spin order in nuclear magnetic resonance: separation of exchange and cross-relaxation processes, Journal of the American Chemical Society, 107 (1985) 6440–6446. [Google Scholar]
- [41].Miloushev VZ, Bahna F, Ciatto C, Ahlsen G, Honig B, Shapiro L, Palmer AG 3rd, Dynamic properties of a type II cadherin adhesive domain: implications for the mechanism of strand-swapping of classical cadherins, Structure, 16 (2008) 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wu C, Wynne SA, Thomas NE, Uhlemann EM, Tate CG, Henzler-Wildman KA, Identification of an Alternating-Access Dynamics Mutant of EmrE with Impaired Transport, Journal of molecular biology, 431 (2019) 2777–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A, NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR, 6 (1995) 277–293. [DOI] [PubMed] [Google Scholar]
- [44].Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K, The program XEASY for computer-supported NMR spectral analysis of biological macromolecules, J. Biomol. NMR, 6 (1995) 1–10. [DOI] [PubMed] [Google Scholar]
- [45].Wishart DS, Sykes BD, The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data, Journal of biomolecular NMR, 4 (1994) 171–180. [DOI] [PubMed] [Google Scholar]
- [46].Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ, Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView, Journal of biomolecular NMR, 18 (2000) 43–48. [DOI] [PubMed] [Google Scholar]
- [47].Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ, Sequence-dependent correction of random coil NMR chemical shifts, Journal of the American Chemical Society, 123 (2001) 2970–2978. [DOI] [PubMed] [Google Scholar]
- [48].Sreerama N, Woody RW, Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set, Anal Biochem, 287 (2000) 252–260. [DOI] [PubMed] [Google Scholar]
- [49].Miles AJ, Ramalli SG, Wallace BA, DichroWeb, a website for calculating protein secondary structure from circular dichroism spectroscopic data, Protein Sci, (2021). [DOI] [PMC free article] [PubMed]
- [50].Schrödinger Release 2021–4: Maestro, in, Schrödinger, LLC, New York, NY, 2021. [Google Scholar]
- [51].Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D, Highly accurate protein structure prediction with AlphaFold, Nature, 596 (2021) 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Zidek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, Kleywegt GJ, Bateman A, Evans R, Pritzel A, Figurnov M, Ronneberger O, Bates R, Kohl SAA, Potapenko A, Ballard AJ, Romera-Paredes B, Nikolov S, Jain R, Clancy E, Reiman D, Petersen S, Senior AW, Kavukcuoglu K, Birney E, Kohli P, Jumper J, Hassabis D, Highly accurate protein structure prediction for the human proteome, Nature, 596 (2021) 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ, Prokaryotic ubiquitin-like protein pup is intrinsically disordered, J Mol Biol, 392 (2009) 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nowicka U, Hoffman M, Randles L, Shi X, Khavrutskii L, Stefanisko K, Tarasova NI, Darwin KH, Walters KJ, Mycobacterium tuberculosis copper-regulated protein SocB is an intrinsically disordered protein that folds upon interaction with a synthetic phospholipid bilayer, Proteins, (2016) 193–200. [DOI] [PMC free article] [PubMed]
- [55].Buel GR, Chen X, Chari R, O’Neill MJ, Ebelle DL, Jenkins C, Sridharan V, Tarasov SG, Tarasova NI, Andresson T, Walters KJ, Structure of E3 ligase E6AP with a proteasome-binding site provided by substrate receptor hRpn10, Nat Commun, 11 (2020) 1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen X, Walters KJ, Identifying and studying ubiquitin receptors by NMR, Methods Mol. Biol, 832 (2012) 279–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen X, Ebelle DL, Wright BJ, Sridharan V, Hooper E, Walters KJ, Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome, J Mol Biol, 431 (2019) 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Whitmore L, Wallace BA, DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data, Nucleic acids research, 32 (2004) W668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Whitmore L, Wallace BA, Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases, Biopolymers, 89 (2008) 392–400. [DOI] [PubMed] [Google Scholar]
- [60].Kim PW, Sun ZY, Blacklow SC, Wagner G, Eck MJ, A zinc clasp structure tethers Lck to T cell coreceptors CD4 and CD8, Science, 301 (2003) 1725–1728. [DOI] [PubMed] [Google Scholar]
- [61].Walters KJ, Dayie KT, Reece RJ, Ptashne M, Wagner G, Structure and mobility of the PUT3 dimer, Nat Struct Biol, 4 (1997) 744–750. [DOI] [PubMed] [Google Scholar]
- [62].Ehlinger A, Park S, Fahmy A, Lary JW, Cole JL, Finley D, Walters KJ, Conformational dynamics of the Rpt6 ATPase in proteasome assembly and Rpn14 binding, Structure, 21 (2013) 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kanelis V, Donaldson L, Muhandiram DR, Rotin D, Forman-Kay JD, Kay LE, Sequential assignment of proline-rich regions in proteins: application to modular binding domain complexes, Journal of biomolecular NMR, 16 (2000) 253–259. [DOI] [PubMed] [Google Scholar]
- [64].Haar W, Fermandjian S, Vicar J, Blaha K, Fromageot P, 13C-nuclear magnetic resonance study of [85% 13C-enriched proline]thyrotropin releasing factor: 13C-13C vicinal coupling constants and conformation of the proline residue, Proceedings of the National Academy of Sciences of the United States of America, 72 (1975) 4948–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Montelione GT, Wagner G, 2D Chemical exchange NMR spectroscopy by proton-detected heteronuclear correlation, J. Am. Chem. Soc, 111 (1989) 3096–3098. [Google Scholar]
- [66].Hudecek M, Sommermeyer D, Kosasih PL, Silva-Benedict A, Liu L, Rader C, Jensen MC, Riddell SR, The nonsignaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity, Cancer Immunol Res, 3 (2015) 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jensen MC, Riddell SR, Designing chimeric antigen receptors to effectively and safely target tumors, Curr Opin Immunol, 33 (2015) 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Theillet FX, Kalmar L, Tompa P, Han KH, Selenko P, Dunker AK, Daughdrill GW, Uversky VN, The alphabet of intrinsic disorder: I. Act like a Pro: On the abundance and roles of proline residues in intrinsically disordered proteins, Intrinsically Disord Proteins, 1 (2013) e24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kulemzin SV, Kuznetsova VV, Mamonkin M, Taranin AV, Gorchakov AA, Engineering Chimeric Antigen Receptors, Acta Naturae, 9 (2017) 6–14. [PMC free article] [PubMed] [Google Scholar]
- [70].Mateos B, Conrad-Billroth C, Schiavina M, Beier A, Kontaxis G, Konrat R, Felli IC, Pierattelli R, The Ambivalent Role of Proline Residues in an Intrinsically Disordered Protein: From Disorder Promoters to Compaction Facilitators, Journal of molecular biology, 432 (2020) 3093–3111. [DOI] [PubMed] [Google Scholar]
- [71].Thomas KM, Naduthambi D, Zondlo NJ, Electronic control of amide cis-trans isomerism via the aromatic-prolyl interaction, Journal of the American Chemical Society, 128 (2006) 2216–2217. [DOI] [PubMed] [Google Scholar]
- [72].Schmid FX, Prolyl isomerases, Adv Protein Chem, 59 (2001) 243–282. [DOI] [PubMed] [Google Scholar]
- [73].Condotta SA, Downey J, Pardy RD, Valbon SF, Tarrab E, Lamarre A, Divangahi M, Richer MJ, Cyclophilin D Regulates Antiviral CD8(+) T Cell Survival in a Cell-Extrinsic Manner, Immunohorizons, 4 (2020) 217–230. [DOI] [PubMed] [Google Scholar]
- [74].Barberi TJ, Dunkle A, He YW, Racioppi L, Means AR, The prolyl isomerase Pin1 modulates development of CD8+ cDC in mice, PLoS One, 7 (2012) e29808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lu KP, Zhou XZ, The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease, Nature reviews. Molecular cell biology, 8 (2007) 904–916. [DOI] [PubMed] [Google Scholar]
- [76].Ramakrishna S, Highfill SL, Walsh Z, Nguyen SM, Lei H, Shern JF, Qin H, Kraft IL, Stetler-Stevenson M, Yuan CM, Hwang JD, Feng Y, Zhu Z, Dimitrov D, Shah NN, Fry TJ, Modulation of Target Antigen Density Improves CAR T-cell Functionality and Persistence, Clin Cancer Res, 25 (2019) 5329–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Walker AJ, Majzner RG, Zhang L, Wanhainen K, Long AH, Nguyen SM, Lopomo P, Vigny M, Fry TJ, Orentas RJ, Mackall CL, Tumor Antigen and Receptor Densities Regulate Efficacy of a Chimeric Antigen Receptor Targeting Anaplastic Lymphoma Kinase, Mol Ther, 25 (2017) 2189–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Shalabi H, Kraft IL, Wang HW, Yuan CM, Yates B, Delbrook C, Zimbelman JD, Giller R, Stetler-Stevenson M, Jaffe ES, Lee DW, Shern JF, Fry TJ, Shah NN, Sequential loss of tumor surface antigens following chimeric antigen receptor T-cell therapies in diffuse large B-cell lymphoma, Haematologica, 103 (2018) e215–e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem, 25 (2004) 1605–1612. [DOI] [PubMed] [Google Scholar]


