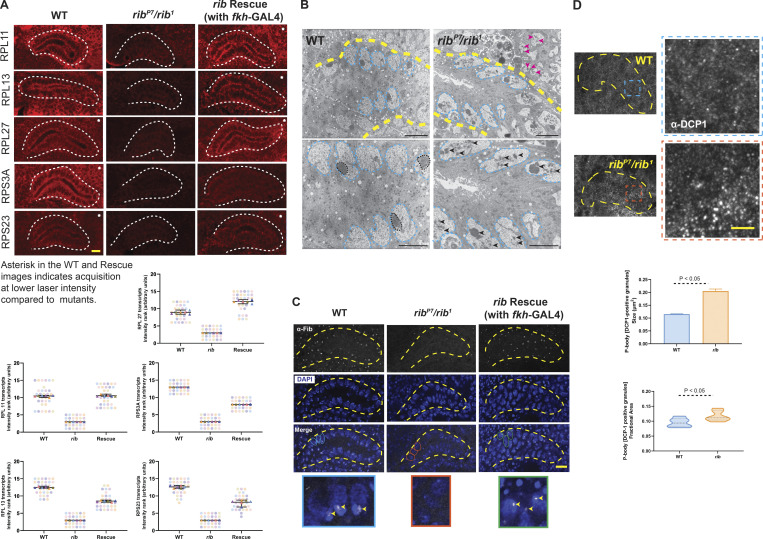
Figure 5.
rib is required for full levels of RPG expression, and rib mutants show defects consistent with ribosome deficiency. (A) Images from FISH of RPGs reveal high-level expression in the WT SG (white outline) and a significant reduction in rib mutants, which is rescued by fkh-GAL4 driven expression of rib (top). SuperPlots showing the scores assigned by six observers ranking RPG transcript signal intensities in 15 blinded samples in the FISH experiments (bottom). Note that the observers consistently ranked what turned out to be the rib-null samples as having the lowest SG signal intensity. Scale bar: 10 µm. See Fig. S7 for DAPI channel. (B) Transmission electron micrographs from sections of embryonic SGs (yellow outline) reveal aberrant nucleolar morphologies in rib mutants (bottom row: slightly higher magnifications). Whereas the nuclear morphologies (cyan outline) are comparable between groups, the electron-dense compaction of nucleolar condensates in WT (black outline) is rarely observed in rib mutants; most rib mutant cells show fragmented or dispersed morphologies characteristic of nucleolar decondensation (black arrowheads). Adjoining mesodermal cell nucleoli also have a fragmented or dispersed morphology (magenta arrowheads) in the rib mutant. SGs from three WT and three rib1/ribP7 embryos were analyzed. Scale bar: 2 µm. (C) Fibrillarin-positive nucleolar punctae observed in WT are lost in rib mutants (yellow outline, SG); fkh-GAL4–driven expression of UAS-rib was sufficient to rescue its localization in SGs and in a subset of surrounding mesoderm-derived cells that express fkh-GAL4. A subset of outlined nuclei (WT, cyan; rib mutant, orange; and rescue, green) in the Merge row is enlarged in the bottom row to highlight the nucleolar Fibrillarin (arrowheads). SGs from four WT, four rib1/ribP7, and four rescue embryos were analyzed. Scale bar: 10 µm. (D) P-body granules (α-DCP1 staining), indicators of untranslated mRNA accumulation, are larger and relatively more abundant in rib mutants than in WT. Top: SG (yellow outline) and the enlarged region of interest (blue/orange boxes) are shown. Scale bar: 2 µm. Bottom: Quantitative analysis showed a significant increase in the P-body granule size in rib SG cells (0.205 ± 0.008 µm2) compared with WT (0.115 ± 0.001 µm2; unpaired t test with two-tailed P value <0.001; mean ± SEM; n = 6 SGs/group). Quantitative analysis of P-body granule fractional area also showed a significant increase in their relative abundance in rib SG cells (0.117 ± 0.007) compared with WT (0.0951 ± 0.006; unpaired t test with two-tailed P value 0.05; median and quartiles; n = 6 SGs/group).