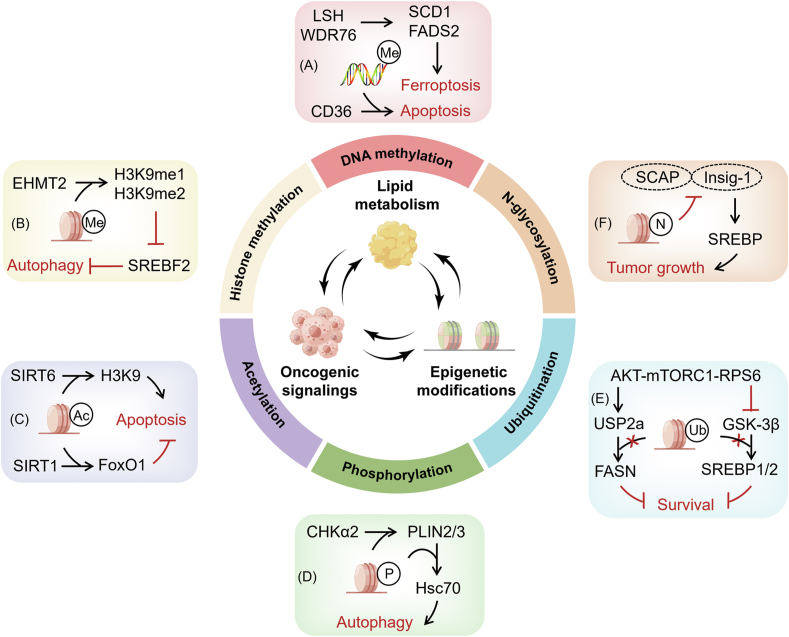
Figure 2.
Crosstalk between lipid metabolism, oncogenic signaling, and epigenetic modifications and their links to metabolic disorders and cancer. Lipid metabolism and the epigenome interact in a bidirectional fashion with genetic and molecular drivers that regulate cancer. An in-depth understanding of the interplay between molecular drivers, metabolic reprogramming, and epigenetic modifications in cancer will clarify their relationships and contribute to the development of effective cancer therapies. (A) Hypermethylation of CD36 leads to apoptosis in lung cancer cells. Increased expression of FADS2 and SCD1 caused by DNA methylation and histone modifications blocks ferroptosis. (B) EHMT2 activates H3K9me1 and H3K9me2, upregulates the transcription of SREBF2, and then triggers autophagy. (C) Deletion of SIRT6 increases promoter H3K9 acetylation levels, resulting in enhanced apoptosis sensitivity. NNMT improves SIRT1 stability while reducing FoxO1 acetylation, lowering the susceptibility of apoptosis. (D) CHK2 phosphorylates PLIN2/3 dissociates from LDs and is degraded by Hsc70-mediated autophagy. (E) The Akt-mTORC1-RPS6 pathway inhibits FASN ubiquitination by upregulating the USP2a de-ubiquitinase and suppresses ubiquitination of SREBP1 and SREBP2 by repressing GSK-3β, thereby promoting hepatocellular carcinoma survival. (F) Glycosylation stabilizes SCAP and decreases its interaction with Insig-1, promoting glioblastoma growth.