Figure 1. Representation of the 3 major bacterial pathways for nitrate reduction to nitrite.
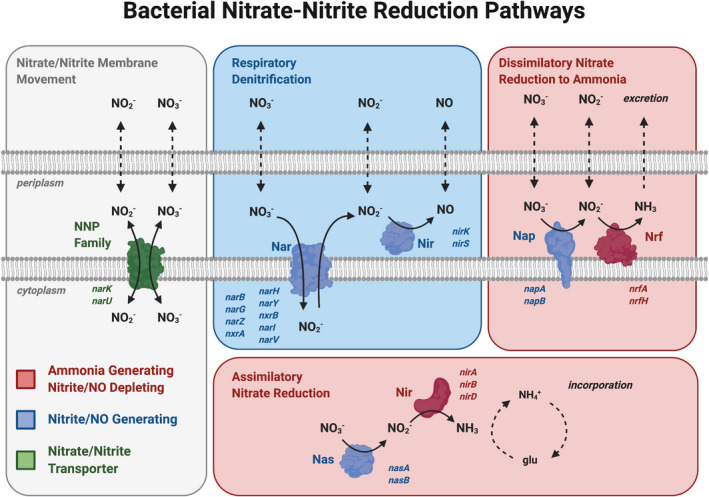
(Adapted from Koch et al 3 and Sparacino‐Watkins et al 15 and created with BioRender.com). Pathways, proteins and genes are color coded. Blue genes and proteins indicate NO2 or NO‐generating enzymes, whereas red indicates nitrite depletion to NH3. Nitrite and nitrate move through the plasma membrane through NNP (nitrate/nitrite transporter ) (in green), coded by the gene narK. The blue box contains the RD pathway that increases nitrite and NO availability through the nitrate reductase Nar (coded by genes narB, narG, narZ, nxrA, narH, narY, nxrB, narI, narV) and nitrite reductase Nir (coded by genes nirK and nirS), respectively, and thus characterized as a nitrite/NO‐generating pathway. Red boxes contain the DNRA and ANR pathways characterized as nitrite‐depleting pathways. Following nitrite generation by nitrate reductases Nap (coded by genes napA and napB) and Nas (coded by genes nasA and nasB), nitrite is depleted by the NH3‐producing enzyme proteins Nir (coded by the genes nirA, nirB, and nirD), and Nrf (coded by the genes nrfA and nrfB). Dotted arrows represent the diffusion of the nitrate, nitrite, nitric oxide, and ammonia in and out of the cell. ANR indicates assimilatory nitrate reduction; DNRA, dissimilatory nitrate reduction to ammonia; glu, glutamine; Nar, Nap, Nas, nitrate reductase; NH3, ammonia; NH4+, ammonium; Nir, Nrf, nitrite reductase; NO2, nitrite; NO3, nitrate; NNP, nitrate/nitrite transporter; and RD, respiratory denitrification.
