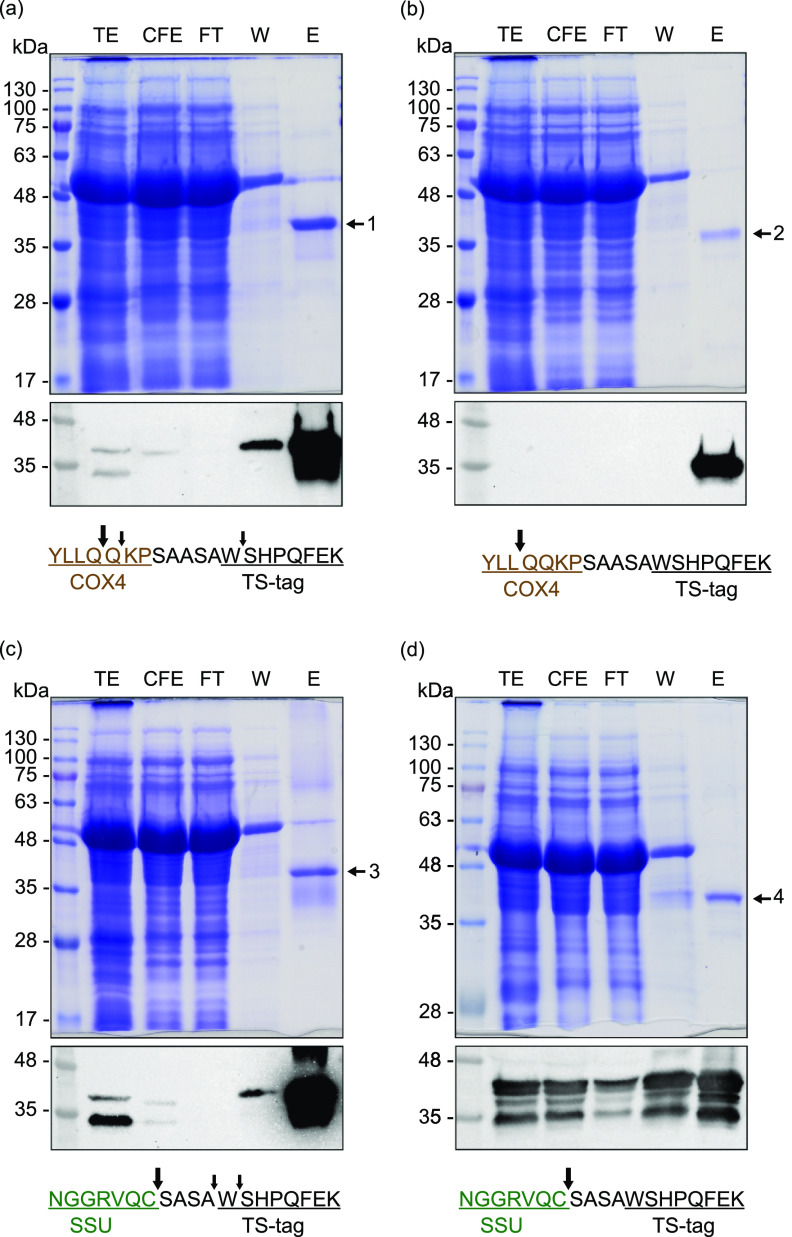
FIG 4.
Purification of soluble NifB proteins from tobacco leaf mitochondria and chloroplasts. Analysis of STAC purification fractions was performed for mitochondrial NifBMi (a), mitochondrial NifBMt (b), chloroplast NifBMi (c), and chloroplast NifBMa (d). Each panel shows the analysis of a representative purification process using Coomassie blue-stained SDS-PAGE and immunoblot analysis with Streptactin-HRP to detect TS-NifB variants. TE, total extract; CFE, cell extract; FT, flowthrough fraction; W, wash fraction; E, elution fraction. Bands 1 to 4 were analyzed by peptide mass fingerprinting and identified as NifBMi (D5VRM1, 55% sequence coverage), NifBMt (O27899, 70% sequence coverage), NifBMi (D5VRM1, 27% sequence coverage), and NifBMa (Q8TIF7, 41% sequence coverage), respectively. Significance peptide mass fingerprint thresholds, P < 0.05. The processing sites for COX4 (mitochondria) and SSU (chloroplast) targeting peptides, as determined by N-terminal sequencing, are shown below each panel. Cleavage sites are indicated by arrows. The dominant processing site is indicated by a larger arrow.