FIG 6.
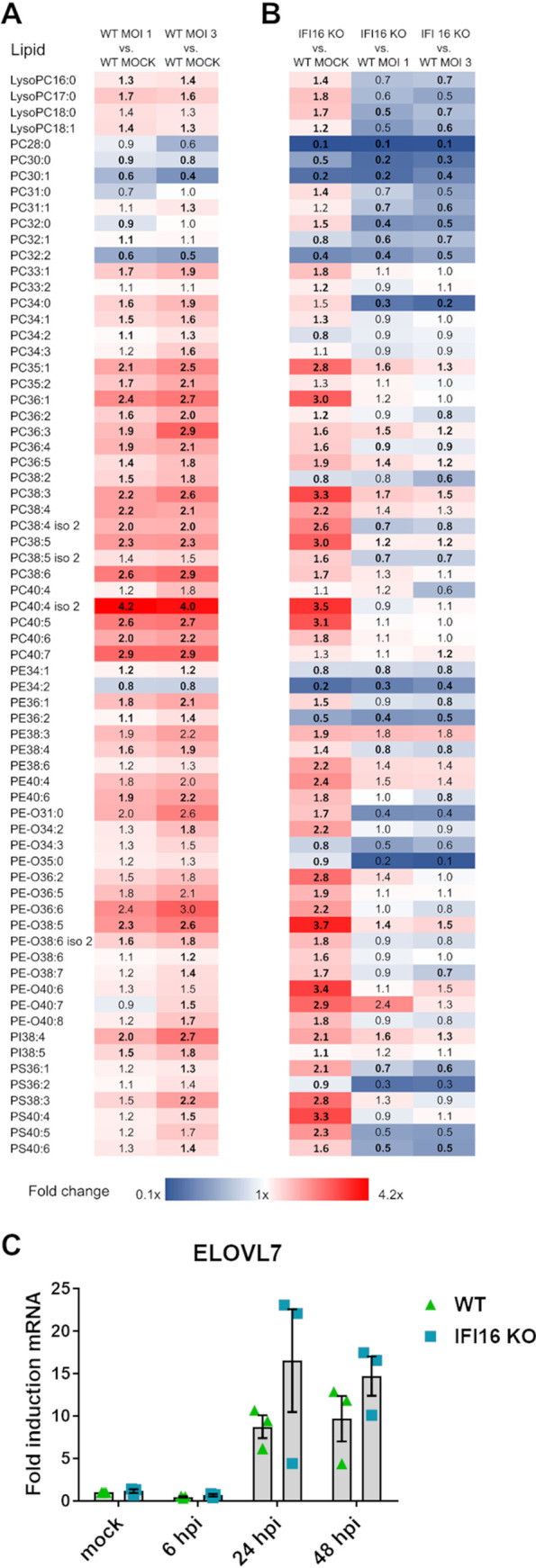
Lipidomic comparison of WT and IFI16 KO HFFs. WT and IFI16 KO HFF cells were infected with HCMV (MOI of 1 or 3) or left uninfected (mock) and maintained in serum-free medium. At 96 hpi, total lipids from cells were extracted and analyzed by LC-MS/MS. The fold changes of lipid species peak area are visualized as a heat map showing the levels of upregulated (red, fold change > 1) and downregulated (blue, fold change < 1) glycerophospholipids. Boldface shows statistically significant values (Mann-Whitney unpaired test, P < 0.05). PC, glycerophosphocholine; PE, glycerophosphoethanolamine; PE-O, ether analogue of glycerophosphoethanolamine; PS, glycerophosphoserine, PI, glycerophosphoinositol. n = 2 independent determinations. (A) Infected WT (MOI of 1 [left] and 3 [right]) versus uninfected HFFs. (B) IFI16 KO versus WT HFFs, mock (left), and HCMV-infected IFI16 KO versus HCMV-infected WT HFFs (MOI of 1 [middle] and 3 [right]). (C) WT and IFI16 KO HFFs were infected with HCMV (MOI, 1), and at 6, 24, and 48 hpi, total RNA was isolated and subjected to RT-qPCR to measure mRNA expression levels of the fatty acid elongase 7 (ELOVL7). Values were normalized to the housekeeping gene GAPDH mRNA and plotted as fold induction relative to WT mock-infected cells (set at 1). Bars show means and SEM from three independent experiments (not significant; two-way ANOVA followed by Bonferroni’s posttests, for comparison of WT versus IFI16 KO cells).
