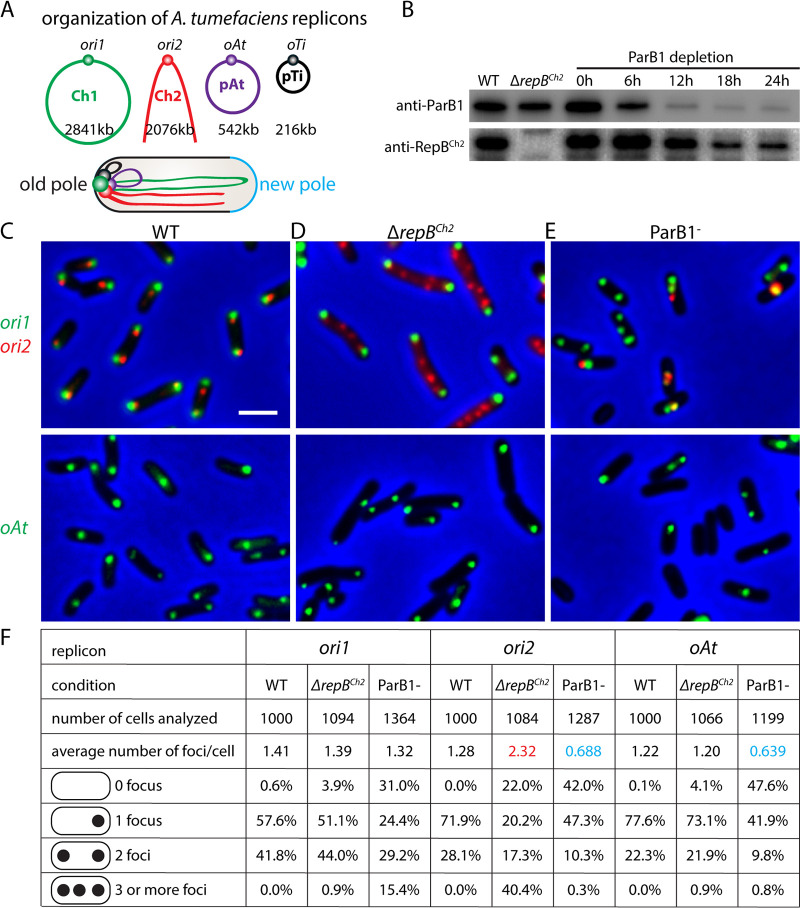
FIG 1.
Centromeric proteins ParB1 and RepBCh2 are important for genome maintenance. (A) Schematic model of the multipartite genome of A. tumefaciens and its cellular organization. In a newborn cell, the four origins are clustered together at the old cell pole, opposite the new pole (light blue). The arms of Ch1 and Ch2 are aligned along the cell length. (B) Immunoblot analysis of ParB1 depletion time course matching microscopy experiments. The levels of ParB1 and RepBCh2 are shown. ParB1 depletion strain (AtWX192) (11) contains PtraI-riboswitch-parB1 at the tetRA locus and parB1 deleted from the endogenous locus. The presence of inducers 1 μM acylhomoserine lactone (AHL) and 2 mM theophylline allows expression of ParB1. Cells were first grown in ATGN solid medium or liquid medium containing inducers. To deplete ParB1, cells were washed in ATGN medium 4 times and then diluted into fresh ATGN without inducers. Cultures were diluted before their optical density at 600 nm (OD600) reached 0.6 to prevent cells from entering stationary phase. Samples were taken at the indicated time points. (C to E) The localization of origins in the wild type (top, AtWX356; bottom, AtWX359) (C) or ΔrepBCh2 (top, AtWX402; bottom, AtWX500) mutant (D) or after 24 h of ParB1 depletion (top, AtWX496; bottom, AtWX498) (E). The origins are labeled using mcherry-parBP1-parSP1 at a position 50 kb away from ori1 (green, top) or ygfp-parBpMT1-parSpMT1 57 kb away from ori2 (red, top) or 11 kb away from oAt (green, bottom). Images of full time course of ParB1 depletion can be found in Fig. S1. Pseudocolors were assigned as indicated. Scale bar represents 2 μm. (F) Analysis of origin number in WT or ΔrepBCh2 strain or ParB1 depletion for 24 h. Images were analyzed using MetaMorph software. Red and blue colors highlight numbers that are higher or lower, respectively, than those for the WT.