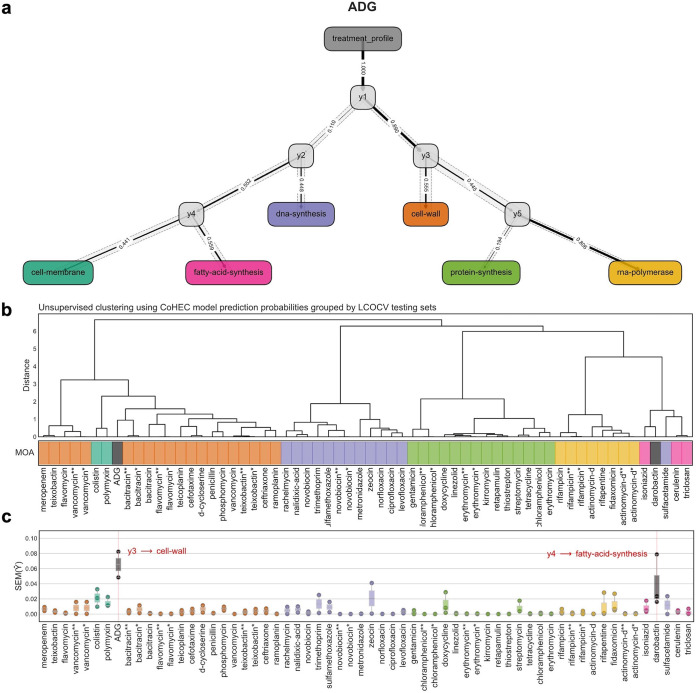
FIG 5.
Predictive modeling based on compounds with known modes of action. (a) CoHEC model decision graph for ADG representing MOA predictions. Prediction paths where each terminal colored node depicts an MOA, each internal gray node represents a submodel decision point, the solid line edge width corresponds to the probability according to the model for the respective path, and the dotted opaque path represents standard errors along the decision path. (b) Unsupervised hierarchical clustering of held-out test set prediction probabilities for each compound unobserved by the model. (c) Standard error profiles for each of the submodel predictions for the held-out test compounds. ADG and darobactin are flagged with novel activity by their high standard error profiles and uncertainty with respect to the model.