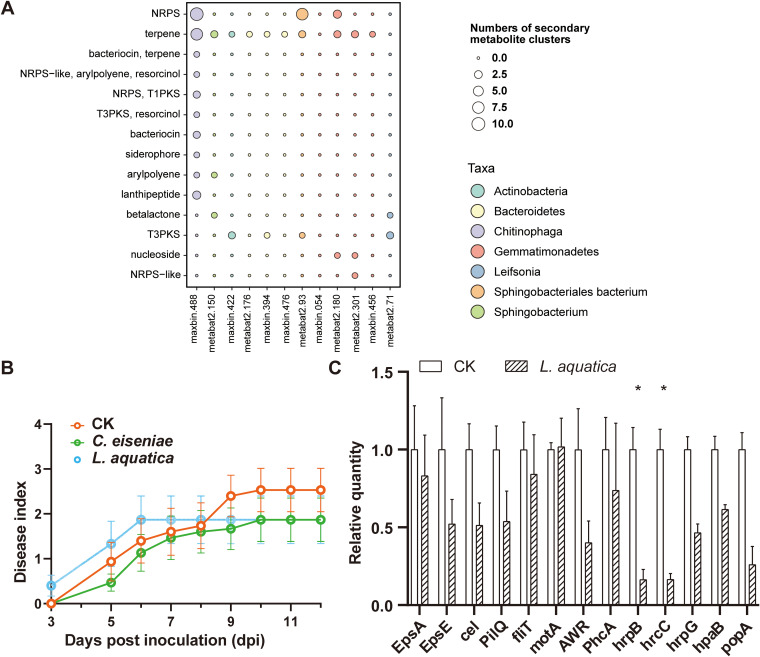
FIG 4.
Bacteria enriched in healthy soil suppress bacterial wilt through multiple strategies. (A) Secondary-metabolite analysis of bins enriched in healthy soil. Type and number of biosynthetic gene clusters (BGCs). Taxonomic annotation of bins is denoted by the color on the right. (B) Progression of bacterial wilt on Moneymaker tomatoes preinoculated with potentially beneficial bacteria in sterilized nursery soil. Each dot represents the mean disease index of 15 tomatoes from three independent experiments, and each error bar indicates the standard error of the mean (SEM; n = 15). (C) Relative quantification using the quantitative reverse transcription-PCR of disease-related genes of Rs treated with the metabolites of L. aquatica. EPS-related genes: epsA and epsE. Drug-proton antiporter gene: cel. Motility-related genes: pilQ, fliT, and motA. T3SS-related genes: phcA, hrpB, hrcC, hrpG, hapB, awr, and popA. The relative absolute abundances of these genes were normalized to the 16S rRNA gene. The relative quantity of each gene was normalized to the control. Each bar height indicates the mean abundance of a gene, and each error bar indicates SEM (n = 4). The level of significance was calculated using two-way ANOVA with Bonferroni’s correction (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).