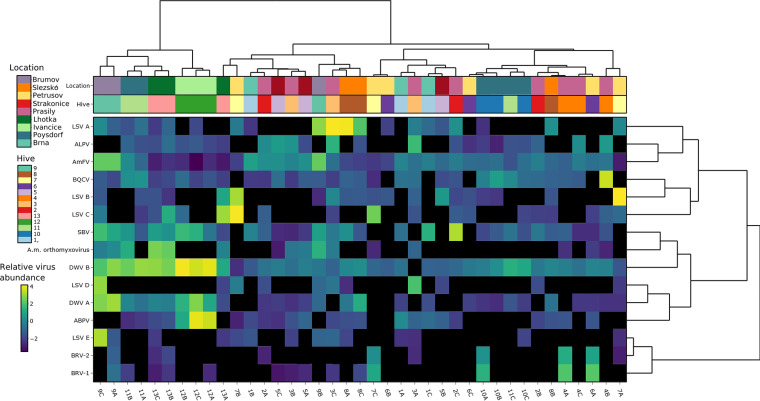
FIG 3.
Diversity of viruses infecting honey bees in healthy bee colonies from Czechia. Relative abundances (viruses per 1 million sequencing reads, calculated from reference genome coverage [see Materials and Methods]) are shown on a log10 scale. Samples (columns) and viruses (rows) are clustered by Ward’s minimum variance method algorithm and seriated by optimal leaf ordering. ABPV, acute bee paralysis virus; ALPV, aphid lethal paralysis virus; AmFV, Apis mellifera filamentous virus; BRV, bee rhabdovirus; BQCV, black queen cell virus; DWV, deformed wing virus; SBV, Sacbrood virus; LSV, Lake Sinai virus.